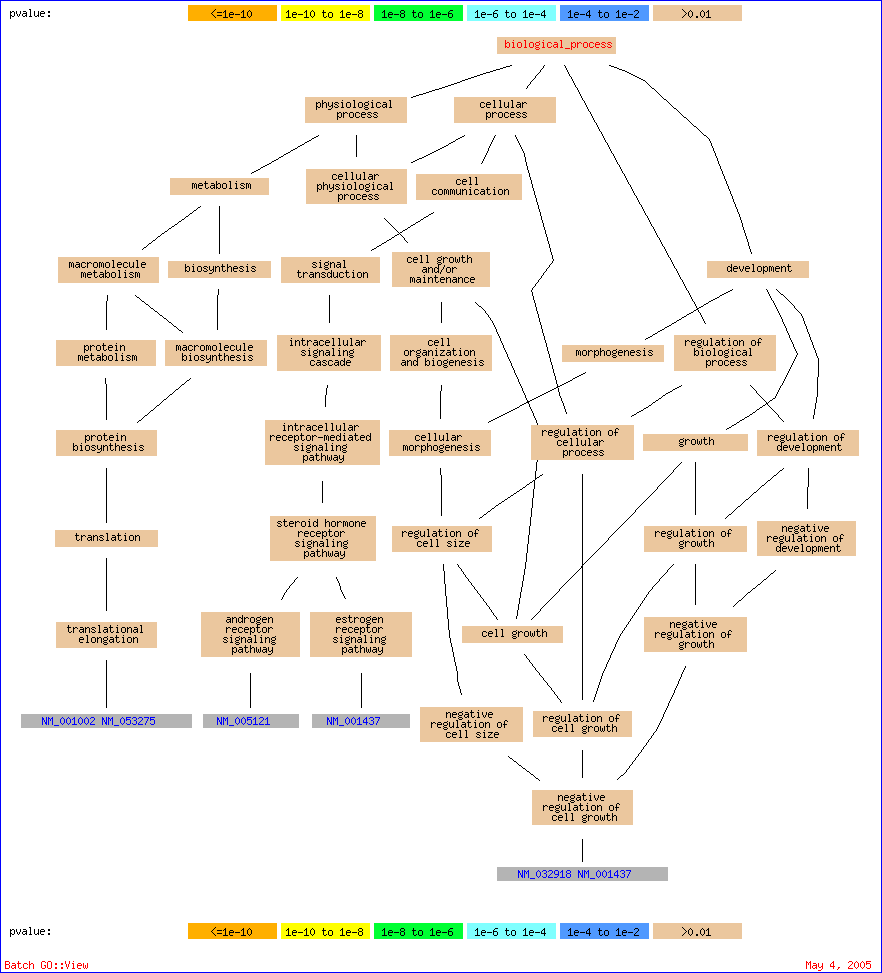
| Gene Ontology term | Cluster frequency | Genome frequency of use | Corrected P-value | FDR | False Positives | Genes annotated to the term |
|---|
| translational elongation | 2 out of 23 genes, 8.7% | 46 out of 30000 genes, 0.2% | 0.06613 | 10.00% | 0.10 | NM_001002, NM_053275 |
| negative regulation of cell growth | 2 out of 23 genes, 8.7% | 52 out of 30000 genes, 0.2% | 0.08448 | 7.00% | 0.14 | NM_032918, NM_001437 |
| negative regulation of cell size | 2 out of 23 genes, 8.7% | 52 out of 30000 genes, 0.2% | 0.08448 | 4.67% | 0.14 | NM_032918, NM_001437 |
| negative regulation of growth | 2 out of 23 genes, 8.7% | 54 out of 30000 genes, 0.2% | 0.09109 | 3.50% | 0.14 | NM_032918, NM_001437 |
| steroid hormone receptor signaling pathway | 2 out of 23 genes, 8.7% | 60 out of 30000 genes, 0.2% | 0.11235 | 3.20% | 0.16 | NM_001437, NM_005121 |
| intracellular receptor-mediated signaling pathway | 2 out of 23 genes, 8.7% | 62 out of 30000 genes, 0.2% | 0.11992 | 3.00% | 0.18 | NM_001437, NM_005121 |
| negative regulation of development | 2 out of 23 genes, 8.7% | 82 out of 30000 genes, 0.3% | 0.20866 | 4.00% | 0.28 | NM_032918, NM_001437 |
| signal transduction | 10 out of 23 genes, 43.5% | 5490 out of 30000 genes, 18.3% | 0.53835 | 10.75% | 0.86 | NM_016602, NM_001437, NM_001345, NM_001459, NM_014326, NM_018184, NM_032918, NM_005314, NM_004767, NM_005121 |
| intracellular signaling cascade | 6 out of 23 genes, 26.1% | 2164 out of 30000 genes, 7.2% | 0.56254 | 10.44% | 0.94 | NM_018184, NM_032918, NM_001437, NM_001345, NM_014326, NM_005121 |
| cell proliferation | 6 out of 23 genes, 26.1% | 2235 out of 30000 genes, 7.4% | 0.65887 | 10.20% | 1.02 | NM_004188, NM_032918, NM_005314, NM_003644, NM_001459, NM_005862 |
| regulation of cell growth | 2 out of 23 genes, 8.7% | 218 out of 30000 genes, 0.7% | 1 | 19.27% | 2.12 | NM_032918, NM_001437 |
| regulation of growth | 2 out of 23 genes, 8.7% | 222 out of 30000 genes, 0.7% | 1 | 17.83% | 2.14 | NM_032918, NM_001437 |
| regulation of cell size | 2 out of 23 genes, 8.7% | 284 out of 30000 genes, 0.9% | 1 | 26.15% | 3.40 | NM_032918, NM_001437 |
| cell growth | 2 out of 23 genes, 8.7% | 284 out of 30000 genes, 0.9% | 1 | 24.29% | 3.40 | NM_032918, NM_001437 |
| translation | 2 out of 23 genes, 8.7% | 297 out of 30000 genes, 1.0% | 1 | 23.87% | 3.58 | NM_001002, NM_053275 |
| regulation of development | 2 out of 23 genes, 8.7% | 304 out of 30000 genes, 1.0% | 1 | 22.75% | 3.64 | NM_032918, NM_001437 |
| G-protein coupled receptor protein signaling pathway | 4 out of 23 genes, 17.4% | 1453 out of 30000 genes, 4.8% | 1 | 22.59% | 3.84 | NM_016602, NM_005314, NM_001345, NM_004767 |
| cell communication | 10 out of 23 genes, 43.5% | 7026 out of 30000 genes, 23.4% | 1 | 23.22% | 4.18 | NM_016602, NM_001437, NM_001345, NM_001459, NM_014326, NM_018184, NM_032918, NM_005314, NM_004767, NM_005121 |
| growth | 2 out of 23 genes, 8.7% | 338 out of 30000 genes, 1.1% | 1 | 22.32% | 4.24 | NM_032918, NM_001437 |
| cellular morphogenesis | 2 out of 23 genes, 8.7% | 356 out of 30000 genes, 1.2% | 1 | 22.90% | 4.58 | NM_032918, NM_001437 |
| cellular process | 15 out of 23 genes, 65.2% | 13216 out of 30000 genes, 44.1% | 1 | 23.71% | 4.98 | NM_004188, NM_016602, NM_001437, NM_001345, NM_001459, NM_014326, NM_006922, NM_018184, NM_053286, NM_032918, NM_005314, NM_003644, NM_004767, NM_005862, NM_005121 |
| protein biosynthesis | 3 out of 23 genes, 13.0% | 981 out of 30000 genes, 3.3% | 1 | 24.27% | 5.34 | NM_001002, NM_053275, NM_001007 |
| mitotic cell cycle | 2 out of 23 genes, 8.7% | 406 out of 30000 genes, 1.4% | 1 | 23.74% | 5.46 | NM_004188, NM_005862 |
| regulation of cellular process | 4 out of 23 genes, 17.4% | 1806 out of 30000 genes, 6.0% | 1 | 26.42% | 6.34 | NM_032918, NM_001437, NM_001459, NM_014326 |
| small GTPase mediated signal transduction | 2 out of 23 genes, 8.7% | 472 out of 30000 genes, 1.6% | 1 | 26.56% | 6.64 | NM_018184, NM_032918 |
| regulation of biological process | 8 out of 23 genes, 34.8% | 5761 out of 30000 genes, 19.2% | 1 | 27.92% | 7.26 | NM_004188, NM_001437, NM_001459, NM_014326, NM_153715, NM_032918, NM_014352, NM_005121 |
| cell growth and/or maintenance | 9 out of 23 genes, 39.1% | 7257 out of 30000 genes, 24.2% | 1 | 34.44% | 9.30 | NM_004188, NM_006922, NM_053286, NM_032918, NM_005314, NM_003644, NM_001437, NM_001459, NM_005862 |
| regulation of cell proliferation | 2 out of 23 genes, 8.7% | 622 out of 30000 genes, 2.1% | 1 | 33.50% | 9.38 | NM_032918, NM_001459 |
| macromolecule biosynthesis | 3 out of 23 genes, 13.0% | 1349 out of 30000 genes, 4.5% | 1 | 32.76% | 9.50 | NM_001002, NM_053275, NM_001007 |
| cellular physiological process | 10 out of 23 genes, 43.5% | 8511 out of 30000 genes, 28.4% | 1 | 33.40% | 10.02 | NM_004188, NM_001437, NM_001459, NM_014326, NM_006922, NM_053286, NM_032918, NM_005314, NM_003644, NM_005862 |
| cell cycle | 3 out of 23 genes, 13.0% | 1499 out of 30000 genes, 5.0% | 1 | 34.65% | 10.74 | NM_004188, NM_003644, NM_005862 |
| cell surface receptor linked signal transduction | 4 out of 23 genes, 17.4% | 2387 out of 30000 genes, 8.0% | 1 | 34.38% | 11.00 | NM_016602, NM_005314, NM_001345, NM_004767 |
| regulation of transcription, DNA-dependent | 5 out of 23 genes, 21.7% | 3473 out of 30000 genes, 11.6% | 1 | 36.67% | 12.10 | NM_004188, NM_014352, NM_001437, NM_153715, NM_005121 |
| regulation of transcription | 5 out of 23 genes, 21.7% | 3571 out of 30000 genes, 11.9% | 1 | 36.88% | 12.54 | NM_004188, NM_014352, NM_001437, NM_153715, NM_005121 |
| transcription, DNA-dependent | 5 out of 23 genes, 21.7% | 3597 out of 30000 genes, 12.0% | 1 | 36.80% | 12.88 | NM_004188, NM_014352, NM_001437, NM_153715, NM_005121 |
| regulation of nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 5 out of 23 genes, 21.7% | 3621 out of 30000 genes, 12.1% | 1 | 36.44% | 13.12 | NM_004188, NM_014352, NM_001437, NM_153715, NM_005121 |
| transcription | 5 out of 23 genes, 21.7% | 3761 out of 30000 genes, 12.5% | 1 | 37.84% | 14.00 | NM_004188, NM_014352, NM_001437, NM_153715, NM_005121 |
| regulation of metabolism | 5 out of 23 genes, 21.7% | 3841 out of 30000 genes, 12.8% | 1 | 38.95% | 14.80 | NM_004188, NM_014352, NM_001437, NM_153715, NM_005121 |
| transcription from Pol II promoter | 2 out of 23 genes, 8.7% | 954 out of 30000 genes, 3.2% | 1 | 38.41% | 14.98 | NM_004188, NM_005121 |
| biosynthesis | 3 out of 23 genes, 13.0% | 1896 out of 30000 genes, 6.3% | 1 | 39.90% | 15.96 | NM_001002, NM_053275, NM_001007 |
| regulation of physiological process | 5 out of 23 genes, 21.7% | 3987 out of 30000 genes, 13.3% | 1 | 40.20% | 16.48 | NM_004188, NM_014352, NM_001437, NM_153715, NM_005121 |
| biological_process unknown | 2 out of 23 genes, 8.7% | 1337 out of 30000 genes, 4.5% | 1 | 47.05% | 19.76 | NM_001345, NM_004767 |
| morphogenesis | 3 out of 23 genes, 13.0% | 2419 out of 30000 genes, 8.1% | 1 | 48.28% | 20.76 | NM_032918, NM_003644, NM_001437 |
| physiological process | 17 out of 23 genes, 73.9% | 19890 out of 30000 genes, 66.3% | 1 | 48.73% | 21.44 | NM_004188, NM_001002, NM_053275, NM_001212, NM_001437, NM_001459, NM_014326, NM_153715, NM_006922, NM_053286, NM_032918, NM_014352, NM_005314, NM_003644, NM_001007, NM_005862, NM_005121 |
| cell organization and biogenesis | 2 out of 23 genes, 8.7% | 1500 out of 30000 genes, 5.0% | 1 | 51.20% | 23.04 | NM_032918, NM_001437 |
| development | 4 out of 23 genes, 17.4% | 3778 out of 30000 genes, 12.6% | 1 | 51.17% | 23.54 | NM_032918, NM_003644, NM_001437, NM_153715 |
| nucleobase, nucleoside, nucleotide and nucleic acid metabolism | 5 out of 23 genes, 21.7% | 5701 out of 30000 genes, 19.0% | 1 | 58.00% | 27.26 | NM_004188, NM_014352, NM_001437, NM_153715, NM_005121 |
| protein metabolism | 4 out of 23 genes, 17.4% | 5140 out of 30000 genes, 17.1% | 1 | 66.96% | 32.14 | NM_001002, NM_053275, NM_014326, NM_001007 |
| macromolecule metabolism | 4 out of 23 genes, 17.4% | 6096 out of 30000 genes, 20.3% | 1 | 71.18% | 34.88 | NM_001002, NM_053275, NM_014326, NM_001007 |
| metabolism | 9 out of 23 genes, 39.1% | 13294 out of 30000 genes, 44.3% | 1 | 71.32% | 35.66 | NM_004188, NM_001002, NM_053275, NM_014352, NM_001437, NM_014326, NM_001007, NM_153715, NM_005121 |
| response to stimulus | 2 out of 23 genes, 8.7% | 3611 out of 30000 genes, 12.0% | 1 | 72.82% | 37.14 | NM_032918, NM_001212 |
| transport | 2 out of 23 genes, 8.7% | 3640 out of 30000 genes, 12.1% | 1 | 72.35% | 37.62 | NM_006922, NM_053286 |
| unannotated | 2 out of 23 genes, 8.7% | 6361 out of 30000 genes, 21.2% | 1 | 78.34% | 41.52 | NM_052900, NM_024722 |