| Gene Ontology term | Cluster frequency | Genome frequency of use | Corrected P-value | FDR | False Positives | Genes annotated to the term |
|---|
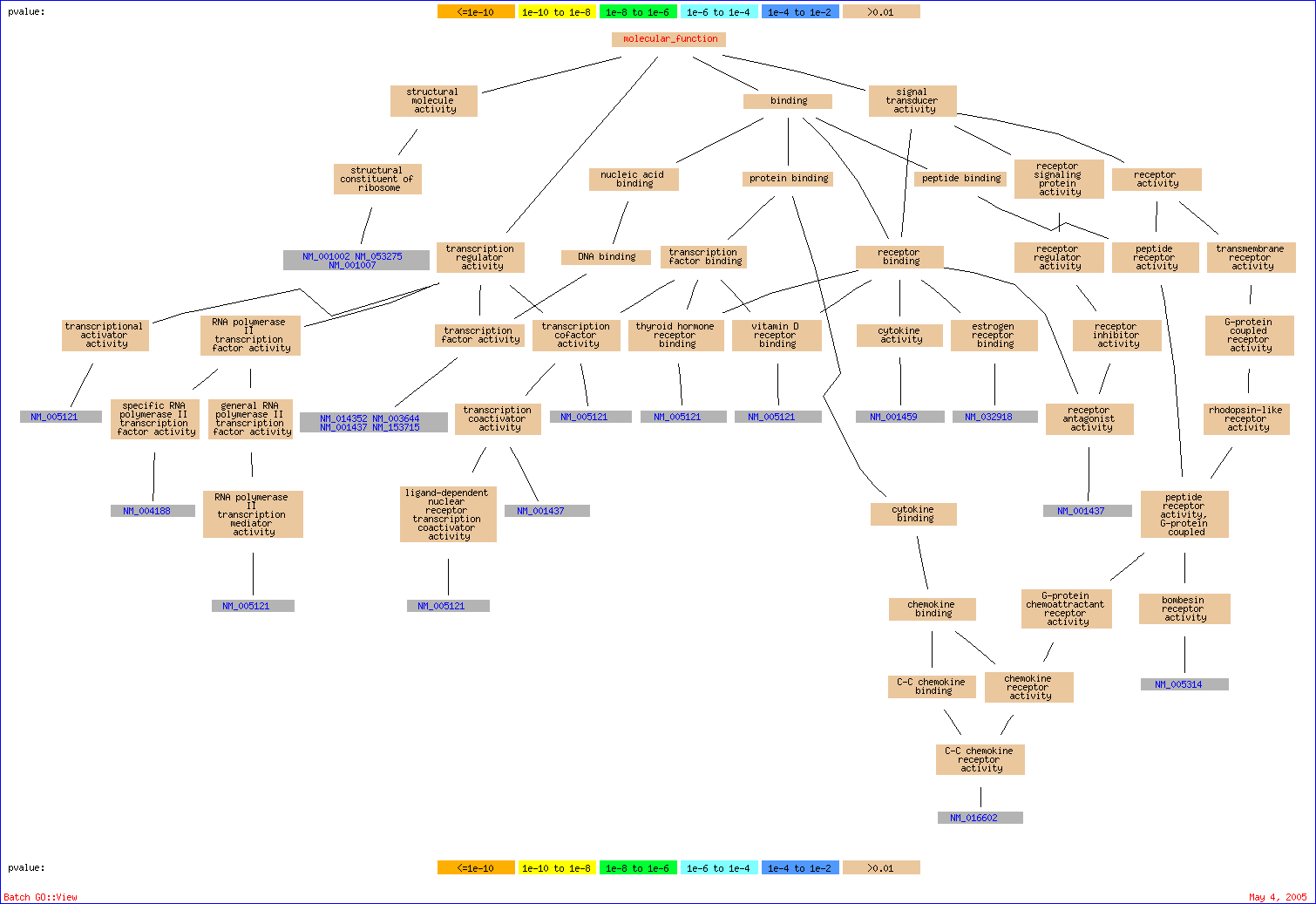
| structural constituent of ribosome | 3 out of 23 genes, 13.0% | 330 out of 30000 genes, 1.1% | 0.18054 | 40.00% | 0.40 | NM_001002, NM_053275, NM_001007 |
| transcription regulator activity | 6 out of 23 genes, 26.1% | 2500 out of 30000 genes, 8.3% | 0.88595 | 50.00% | 1.00 | NM_004188, NM_014352, NM_003644, NM_001437, NM_153715, NM_005121 |
| peptide receptor activity | 2 out of 23 genes, 8.7% | 236 out of 30000 genes, 0.8% | 1 | 46.00% | 1.38 | NM_016602, NM_005314 |
| peptide receptor activity, G-protein coupled | 2 out of 23 genes, 8.7% | 236 out of 30000 genes, 0.8% | 1 | 34.50% | 1.38 | NM_016602, NM_005314 |
| receptor binding | 4 out of 23 genes, 17.4% | 1269 out of 30000 genes, 4.2% | 1 | 28.40% | 1.42 | NM_032918, NM_001437, NM_001459, NM_005121 |
| peptide binding | 2 out of 23 genes, 8.7% | 296 out of 30000 genes, 1.0% | 1 | 29.67% | 1.78 | NM_016602, NM_005314 |
| lipid binding | 2 out of 23 genes, 8.7% | 308 out of 30000 genes, 1.0% | 1 | 25.71% | 1.80 | NM_024722, NM_001345 |
| binding | 17 out of 23 genes, 73.9% | 15390 out of 30000 genes, 51.3% | 1 | 22.50% | 1.80 | NM_004188, NM_001002, NM_053275, NM_016602, NM_001437, NM_001345, NM_001459, NM_014326, NM_153715, NM_018184, NM_032918, NM_014352, NM_005314, NM_003644, NM_024722, NM_001007, NM_005121 |
| rhodopsin-like receptor activity | 3 out of 23 genes, 13.0% | 813 out of 30000 genes, 2.7% | 1 | 20.44% | 1.84 | NM_016602, NM_005314, NM_004767 |
| transcription coactivator activity | 2 out of 23 genes, 8.7% | 348 out of 30000 genes, 1.2% | 1 | 23.60% | 2.36 | NM_001437, NM_005121 |
| RNA binding | 3 out of 23 genes, 13.0% | 980 out of 30000 genes, 3.3% | 1 | 24.91% | 2.74 | NM_001002, NM_053275, NM_001007 |
| G-protein coupled receptor activity | 3 out of 23 genes, 13.0% | 1021 out of 30000 genes, 3.4% | 1 | 25.67% | 3.08 | NM_016602, NM_005314, NM_004767 |
| RNA polymerase II transcription factor activity | 2 out of 23 genes, 8.7% | 450 out of 30000 genes, 1.5% | 1 | 25.38% | 3.30 | NM_004188, NM_005121 |
| transcription factor activity | 4 out of 23 genes, 17.4% | 1880 out of 30000 genes, 6.3% | 1 | 26.29% | 3.68 | NM_014352, NM_003644, NM_001437, NM_153715 |
| nucleic acid binding | 8 out of 23 genes, 34.8% | 5856 out of 30000 genes, 19.5% | 1 | 28.53% | 4.28 | NM_004188, NM_001002, NM_053275, NM_014352, NM_003644, NM_001437, NM_001007, NM_153715 |
| transcription cofactor activity | 2 out of 23 genes, 8.7% | 575 out of 30000 genes, 1.9% | 1 | 28.50% | 4.56 | NM_001437, NM_005121 |
| receptor activity | 5 out of 23 genes, 21.7% | 3042 out of 30000 genes, 10.1% | 1 | 28.47% | 4.84 | NM_016602, NM_005314, NM_001437, NM_004767, NM_005121 |
| structural molecule activity | 3 out of 23 genes, 13.0% | 1322 out of 30000 genes, 4.4% | 1 | 27.67% | 4.98 | NM_001002, NM_053275, NM_001007 |
| GTP binding | 2 out of 23 genes, 8.7% | 622 out of 30000 genes, 2.1% | 1 | 27.89% | 5.30 | NM_018184, NM_032918 |
| signal transducer activity | 7 out of 23 genes, 30.4% | 5101 out of 30000 genes, 17.0% | 1 | 26.70% | 5.34 | NM_032918, NM_016602, NM_005314, NM_001437, NM_001459, NM_004767, NM_005121 |
| guanyl nucleotide binding | 2 out of 23 genes, 8.7% | 638 out of 30000 genes, 2.1% | 1 | 25.81% | 5.42 | NM_018184, NM_032918 |
| transcription factor binding | 2 out of 23 genes, 8.7% | 639 out of 30000 genes, 2.1% | 1 | 25.18% | 5.54 | NM_001437, NM_005121 |
| obsolete molecular function | 2 out of 23 genes, 8.7% | 992 out of 30000 genes, 3.3% | 1 | 36.26% | 8.34 | NM_018184, NM_032918 |
| transmembrane receptor activity | 3 out of 23 genes, 13.0% | 2037 out of 30000 genes, 6.8% | 1 | 39.58% | 9.50 | NM_016602, NM_005314, NM_004767 |
| phosphotransferase activity, alcohol group as acceptor | 2 out of 23 genes, 8.7% | 1549 out of 30000 genes, 5.2% | 1 | 55.52% | 13.88 | NM_001345, NM_014326 |
| DNA binding | 4 out of 23 genes, 17.4% | 3973 out of 30000 genes, 13.2% | 1 | 54.77% | 14.24 | NM_014352, NM_003644, NM_001437, NM_153715 |
| kinase activity | 2 out of 23 genes, 8.7% | 1763 out of 30000 genes, 5.9% | 1 | 53.85% | 14.54 | NM_001345, NM_014326 |
| protein binding | 4 out of 23 genes, 17.4% | 4262 out of 30000 genes, 14.2% | 1 | 52.43% | 14.68 | NM_016602, NM_001437, NM_014326, NM_005121 |
| transferase activity, transferring phosphorus-containing groups | 2 out of 23 genes, 8.7% | 1854 out of 30000 genes, 6.2% | 1 | 52.14% | 15.12 | NM_001345, NM_014326 |
| purine nucleotide binding | 3 out of 23 genes, 13.0% | 3158 out of 30000 genes, 10.5% | 1 | 53.07% | 15.92 | NM_018184, NM_032918, NM_014326 |
| nucleotide binding | 3 out of 23 genes, 13.0% | 3194 out of 30000 genes, 10.6% | 1 | 52.45% | 16.26 | NM_018184, NM_032918, NM_014326 |
| transferase activity | 2 out of 23 genes, 8.7% | 3074 out of 30000 genes, 10.2% | 1 | 65.81% | 21.06 | NM_001345, NM_014326 |
| transporter activity | 2 out of 23 genes, 8.7% | 3201 out of 30000 genes, 10.7% | 1 | 66.18% | 21.84 | NM_006922, NM_053286 |
| unannotated | 3 out of 23 genes, 13.0% | 4732 out of 30000 genes, 15.8% | 1 | 65.12% | 22.14 | NM_052900, NM_001212, NM_005862 |
| cation binding | 2 out of 23 genes, 8.7% | 3473 out of 30000 genes, 11.6% | 1 | 64.63% | 22.62 | NM_004188, NM_001345 |
| ion binding | 2 out of 23 genes, 8.7% | 3811 out of 30000 genes, 12.7% | 1 | 64.22% | 23.12 | NM_004188, NM_001345 |
| metal ion binding | 2 out of 23 genes, 8.7% | 3811 out of 30000 genes, 12.7% | 1 | 62.49% | 23.12 | NM_004188, NM_001345 |
| catalytic activity | 2 out of 23 genes, 8.7% | 8533 out of 30000 genes, 28.4% | 1 | 65.58% | 24.92 | NM_001345, NM_014326 |