Annotation results
RNA/H3ac/H3K27me3/HS/RFBR/TR50, 2-state segmentation (including Figure 9a from Nature paper)
|
Gene overlapBed files below contain one line for each gene whose Tx start sits inside the given state. Columns are chr, start, stop, gene ID, fraction of gene sitting inside the segment, strand, and segment ID
|
Enrichment of various elements
Total number of bases or elements overlapping each state. Numbers in parentheses are the fractions of the element in that state. Second set of columns gives enrichment over expected (as a fraction). Enrichment p-values, computed by permuting state labels on the fixed segmentation, are also included, with adjusted p-values using method "BH".Element st0 st1 st0 enr st0 enr p st0 adj p st1 enr st1 enr p st1 adj p ------------------------- -------------------- -------------------- --------- ---------- ---------- --------- ---------- ---------- Num_Gencode_TxStarts 711 (0.262) 1998 (0.738) -0.569 0 0 0.886 0 0 Num_mRNA_TxStarts 980 (0.212) 3635 (0.788) -0.651 0 0 1.014 0 0 Num_Spliced.EST_TxStarts 15771 (0.121) 114949 (0.879) -0.802 0 0 1.249 0 0 EST_Ovr 10619024 (0.558) 8407889 (0.442) -0.083 0.0192 0.02671 0.130 0.0042 0.007074 CpG.UCSC_Isl_Ovr 71486 (0.189) 305944 (0.811) -0.689 0 0 1.073 0 0 RptMskr_Ovr 8051498 (0.609) 5167707 (0.391) 0.000 0.4854 0.4854 -0.000 0.4824 0.4854 RptMskr_DNA_Ovr 529253 (0.651) 283505 (0.349) 0.069 0.0443 0.0525 -0.108 0.0105 0.016 RptMskr_LINE.L1_Ovr 3001664 (0.732) 1096442 (0.268) 0.203 0 0 -0.316 0 0 RptMskr_LINE.L2_Ovr 647739 (0.649) 351056 (0.351) 0.065 0.053 0.06057 -0.101 0.0181 0.02633 RptMskr_LTR_Ovr 1451037 (0.706) 604366 (0.294) 0.159 0.0004 0.0009143 -0.248 0 0 RptMskr_SINE.ALU_Ovr 1519386 (0.401) 2266702 (0.599) -0.341 0 0 0.531 0 0 SimpRep_Ovr 331584 (0.480) 359151 (0.520) -0.212 0.0036 0.0064 0.330 0.0005 0.001067 MSA_strict_Ovr 362948 (0.508) 351053 (0.492) -0.165 0.0364 0.0448 0.257 0.0055 0.0088 MSA_moderate_Ovr 760236 (0.520) 702794 (0.480) -0.147 0.0217 0.02893 0.228 0.0014 0.0028 MSA_loose_Ovr 1927072 (0.553) 1557686 (0.447) -0.092 0.0233 0.02982 0.143 0.0028 0.005271 MSA_non.exonic_Ovr 602029 (0.674) 291703 (0.326) 0.106 0.1511 0.1612 -0.165 0.0901 0.09942
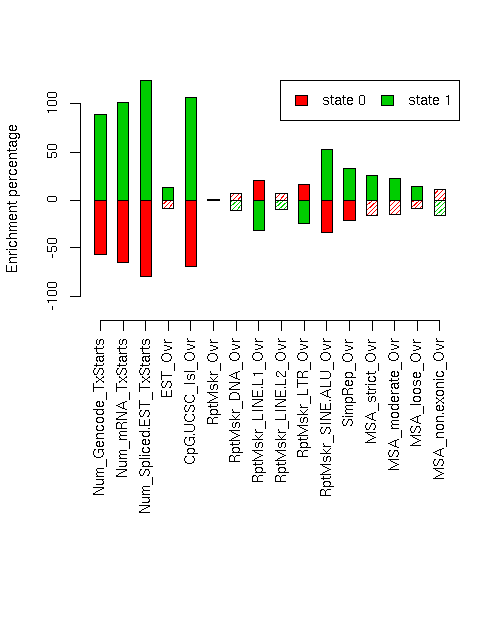
(Postscript version of above - edited version of this is Figure 9b from Nature paper)
Distances of elements to segment boundaries
Average and median absolute value over all boundaries.
Statistic avg(|d|) med(|d|) ------------------------- ------------ ------------ Gencode_5P_Dist_Known 1549915.68 17839.0 Gencode_3P_Dist_Known 1551649.22 18050.0 Gencode_5P_Dist_Putative 2788590.76 91513.0 Gencode_3P_Dist_Putative 2787346.84 90609.0 Gencode_5P_Dist_Pseudo 1222619.30 84648.0 Gencode_3P_Dist_Pseudo 1222361.13 83365.0 Gencode_TxStop_Dist 384221.54 10315.0 Gencode_TxStart_Dist 385316.57 10317.0 Gencode_5P.UTR_Dist 1559102.17 17806.0 Gencode_1stExon_Dist 386063.50 10642.0 KnownGene_3P_Dist 89099.92 35994.0 KnownGene_5P_Dist 83937.66 32431.0 Ensemble_5P_Dist 82615.47 31060.0 Refseq_5P_Dist 90723.30 41872.0 mRNA_5P_Dist 47249.02 14097.0 mRNA_3P_Dist 47207.45 19327.0 Spliced.EST_5P_Dist 15778.93 4105.0 Spliced.EST_3P_Dist 13831.59 3487.0 CpG.UCSC_Isl_Dist 95964.00 39769.0 CpG.AL_Isl_Dist 739635.39 24645.0 Nhgri_Chip.GM_Dist 716353.58 16745.0 UWDHS.CAP.GM_Dist 449428.27 13884.0 UWDHS.QCP.GM_Dist 20111490.65 73614.0 UWDHS.QCP.HeLa_Dist 20105597.94 55428.5 UVaDnaRep.OriginsPred_Dist 52471.24 39590.0 Sanger_ChipCenters.H3K4me1.GM_Dist 23724.20 9365.0 Sanger_ChipCenters.H3K4me1.HeLa_Dist 406572.24 18749.0 Sanger_ChipCenters.H3K4me2.GM_Dist 30087.13 11191.0 Sanger_ChipCenters.H3K4me2.HeLa_Dist 2114431.12 36461.0 Sanger_ChipCenters.H3K4me3.GM_Dist 41532.65 19554.0 Sanger_ChipCenters.H3K4me3.HeLa_Dist 2805855.52 35779.0 Sanger_ChipCenters.H3ac.GM_Dist 69221.01 28783.0 Sanger_ChipCenters.H3ac.HeLa_Dist 2280139.49 44040.0 Sanger_ChipCenters.H4ac.GM_Dist 56493.01 25661.0 Sanger_ChipCenters.H4ac.HeLa_Dist 1236929.51 42190.0 Uva_DnaRep.Early_Dist 8325831.15 92976.0 Uva_DnaRep.Mid_Dist 5075146.12 38981.0 Uva_DnaRep.Late_Dist 682484.63 66990.0 Uva_DnaRep.PanS_Dist 4595555.56 173276.0 MSA_non.exonic_Dist 6065.76 2885.0