Annotation results
Uva TR50 data, HeLa, 2-state segmentation (state 0 = active!)
|
Gene overlapBed files below contain one line for each gene whose Tx start sits inside the given state. Columns are chr, start, stop, gene ID, fraction of gene sitting inside the segment, strand, and segment ID
|
Enrichment of various elements
Total number of bases or elements overlapping each state. Numbers in parentheses are the fractions of the element in that state. Second set of columns gives enrichment over expected (as a fraction). Enrichment p-values, computed by permuting state labels on the fixed segmentation, are also included, with adjusted p-values using method "BH".Element st0 st1 st0 enr st0 enr p st0 adj p st1 enr st1 enr p st1 adj p ------------------------- -------------------- -------------------- --------- ---------- ---------- --------- ---------- ---------- Num_Gencode_TxStarts 2050 (0.766) 626 (0.234) 0.324 0.0062 0.01804 -0.445 0.0005 0.0032 Num_mRNA_TxStarts 3643 (0.794) 944 (0.206) 0.372 0.0414 0.06309 -0.511 0.0036 0.0128 Num_Spliced.EST_TxStarts 113086 (0.867) 17421 (0.133) 0.497 0.2131 0.31 -0.683 0.0143 0.02805 EST_Ovr 11753917 (0.629) 6940103 (0.371) 0.086 0.0383 0.06309 -0.119 0.0149 0.02805 CpG.UCSC_Isl_Ovr 290627 (0.788) 78423 (0.212) 0.361 0.0103 0.02354 -0.496 0.0002 0.002133 RptMskr_Ovr 7603009 (0.585) 5399303 (0.415) 0.010 0.2874 0.3679 -0.014 0.2293 0.319 RptMskr_DNA_Ovr 466531 (0.581) 336239 (0.419) 0.004 0.4631 0.4631 -0.006 0.4525 0.4631 RptMskr_LINE.L1_Ovr 2033172 (0.502) 2014051 (0.498) -0.132 0.0089 0.02191 0.181 0.001 0.005333 RptMskr_LINE.L2_Ovr 561846 (0.574) 417754 (0.426) -0.009 0.434 0.4629 0.012 0.4134 0.4629 RptMskr_LTR_Ovr 1003425 (0.497) 1014096 (0.503) -0.141 0.0016 0.0064 0.193 0.0001 0.0016 RptMskr_SINE.ALU_Ovr 2763642 (0.744) 950773 (0.256) 0.286 0.0005 0.0032 -0.392 0 0 SimpRep_Ovr 357587 (0.533) 313838 (0.467) -0.080 0.314 0.3865 0.110 0.2523 0.3364 MSA_strict_Ovr 473982 (0.666) 237857 (0.334) 0.150 0.0409 0.06309 -0.207 0.0084 0.02191 MSA_moderate_Ovr 953775 (0.656) 500792 (0.344) 0.133 0.0288 0.0512 -0.183 0.0059 0.01804 MSA_loose_Ovr 2201836 (0.638) 1250368 (0.362) 0.102 0.0145 0.02805 -0.140 0.0015 0.0064 MSA_non.exonic_Ovr 505808 (0.567) 385591 (0.433) -0.020 0.4246 0.4629 0.027 0.4021 0.4629
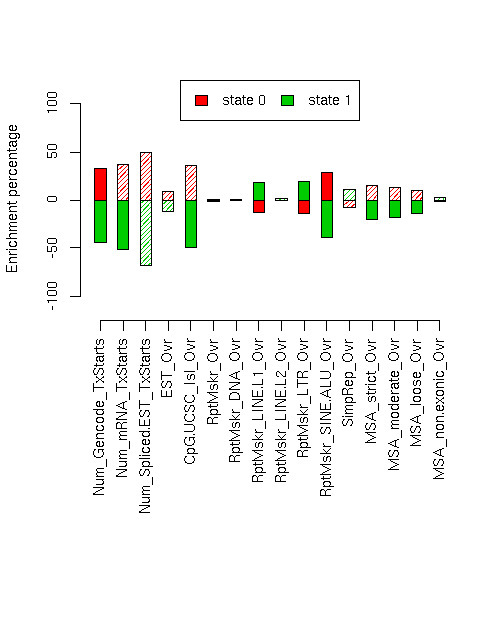
Distances of elements to segment boundaries
Average and median absolute value over all boundaries.
Statistic avg(|d|) med(|d|) ------------------------- ------------ ------------ Gencode_5P_Dist_Known 2532759.42 20406.0 Gencode_3P_Dist_Known 2531035.48 26590.0 Gencode_5P_Dist_Putative 3816149.75 138842.0 Gencode_3P_Dist_Putative 3814031.28 143266.0 Gencode_5P_Dist_Pseudo 2240602.19 121504.0 Gencode_3P_Dist_Pseudo 2240426.35 112402.0 Gencode_TxStop_Dist 545913.72 12385.0 Gencode_TxStart_Dist 548619.48 11939.0 Gencode_5P.UTR_Dist 2545128.63 24432.0 Gencode_1stExon_Dist 549677.47 14346.0 KnownGene_3P_Dist 119788.64 45832.0 KnownGene_5P_Dist 116283.13 39022.0 Ensemble_5P_Dist 114109.74 42397.0 Refseq_5P_Dist 120900.47 47690.0 mRNA_5P_Dist 66462.40 19803.0 mRNA_3P_Dist 64984.62 25107.0 Spliced.EST_5P_Dist 18988.01 4476.0 Spliced.EST_3P_Dist 16486.99 3108.0 CpG.UCSC_Isl_Dist 144064.36 57745.0 CpG.AL_Isl_Dist 1058199.51 27165.0 Nhgri_Chip.GM_Dist 1013955.15 17557.0 UWDHS.CAP.GM_Dist 633119.45 23673.0 UWDHS.QCP.GM_Dist 21250180.09 956447.0 UWDHS.QCP.HeLa_Dist 21241171.03 933743.0 UVaDnaRep.OriginsPred_Dist 61024.95 49197.0 Sanger_ChipCenters.H3K4me1.GM_Dist 30649.32 14590.0 Sanger_ChipCenters.H3K4me1.HeLa_Dist 579849.69 30001.0 Sanger_ChipCenters.H3K4me2.GM_Dist 39441.23 18589.0 Sanger_ChipCenters.H3K4me2.HeLa_Dist 3357107.73 65098.0 Sanger_ChipCenters.H3K4me3.GM_Dist 54383.19 26985.0 Sanger_ChipCenters.H3K4me3.HeLa_Dist 4874746.61 62894.0 Sanger_ChipCenters.H3ac.GM_Dist 81246.82 46538.0 Sanger_ChipCenters.H3ac.HeLa_Dist 3760385.10 84958.0 Sanger_ChipCenters.H4ac.GM_Dist 70065.26 44899.0 Sanger_ChipCenters.H4ac.HeLa_Dist 2097119.84 81286.0 Uva_DnaRep.Early_Dist 9926312.06 149191.5 Uva_DnaRep.Mid_Dist 4089138.76 19698.0 Uva_DnaRep.Late_Dist 765624.61 17349.0 Uva_DnaRep.PanS_Dist 5509666.08 159074.0 MSA_non.exonic_Dist 9165.43 3329.0