Annotation results
SangerChip data, H3K4me1, HeLa, 64kb wavelet smooth data, 2-state segmentation
|
Gene overlapBed files below contain one line for each gene whose Tx start sits inside the given state. Columns are chr, start, stop, gene ID, fraction of gene sitting inside the segment, strand, and segment ID
|
Overlap with various elements
Total number of bases overlapping various elements in each state. Numbers in parentheses are the fractions of the element in that state. Second set of columns gives enrichment over expected (as a fraction). Enrichment p-values, computed by permuting state labels on the fixed segmentation, are also included. The third set of columns gives fraction of each state's segments that overlap the given element.Element st0 st1 st0 enr st0 enr p st1 enr st1 enr p st0 (frac) st1 (frac) ------------------------- -------------------- -------------------- --------- ---------- --------- ---------- ---------- ---------- Gencode_Known_Exon_Ovr 637121 (0.386) 1015518 (0.614) -0.323 0 0.429 0 0.386 0.614 Gencode_Putative_Exon_Ovr 66382 (0.502) 65722 (0.498) -0.118 0.1662 0.157 0.1418 0.502 0.498 Gencode_Pseudo_Exon_Ovr 91641 (0.674) 44391 (0.326) 0.182 0.1973 -0.241 0.1691 0.674 0.326 Gencode_Intron_Ovr 8905184 (0.528) 7951227 (0.472) -0.073 0.0528 0.097 0.0322 0.528 0.472 Gencode_Coding_Exon_Ovr 360993 (0.447) 446386 (0.553) -0.215 0.0116 0.285 0.0069 0.447 0.553 Gencode_5P.UTR_Ovr 226014 (0.355) 410734 (0.645) -0.377 0.0001 0.500 0.0001 0.355 0.645 Gencode_3P.UTR_Ovr 367675 (0.400) 551581 (0.600) -0.298 0 0.395 0.0001 0.400 0.600 Gencode_1stExon_Ovr 305385 (0.386) 485803 (0.614) -0.323 0.0003 0.427 0.0002 0.386 0.614 KnownGene_Intron_Ovr 6763136 (0.500) 6764659 (0.500) -0.123 0.0266 0.163 0.0177 0.500 0.500 KnownGene_Exon_Ovr 454933 (0.402) 676895 (0.598) -0.295 0 0.390 0.0001 0.402 0.598 KnownGene_5P.UTR_Ovr 51438 (0.359) 91705 (0.641) -0.369 0 0.489 0.0001 0.359 0.641 KnownGene_3P.UTR_Ovr 180498 (0.412) 257175 (0.588) -0.276 0.0002 0.366 0.0007 0.412 0.588 EST_Ovr 9957782 (0.512) 9506241 (0.488) -0.102 0.0052 0.135 0.0021 0.512 0.488 Gene.Unit.Ovr 11227003 (0.543) 9457180 (0.457) -0.048 0.128 0.063 0.1022 0.543 0.457 CpG.UCSC_Isl_Ovr 107451 (0.272) 287525 (0.728) -0.523 0 0.692 0 0.272 0.728 CpG.AL_Isl_Ovr 325051 (0.309) 726524 (0.691) -0.458 0 0.606 0.0002 0.309 0.691 RptMskr_Ovr 7746464 (0.577) 5672727 (0.423) 0.013 0.2036 -0.017 0.1721 0.577 0.423 RptMskr_DNA_Ovr 502939 (0.612) 319455 (0.388) 0.073 0.026 -0.097 0.0146 0.612 0.388 RptMskr_LINE_Ovr 3458567 (0.655) 1821657 (0.345) 0.149 0 -0.198 0 0.655 0.345 RptMskr_LINE.L1_Ovr 2821696 (0.676) 1349453 (0.324) 0.187 0 -0.248 0 0.676 0.324 RptMskr_LINE.L2_Ovr 574832 (0.572) 430263 (0.428) 0.004 0.4706 -0.005 0.4656 0.572 0.428 RptMskr_LTR_Ovr 1380777 (0.659) 713542 (0.341) 0.157 0.0003 -0.208 0.0002 0.659 0.341 RptMskr_SINE.ALU_Ovr 1635336 (0.427) 2190980 (0.573) -0.250 0 0.331 0 0.427 0.573 RptMskr_SINE_Ovr 2137283 (0.452) 2587385 (0.548) -0.206 0 0.273 0 0.452 0.548 SimpRep_Ovr 376401 (0.501) 374525 (0.499) -0.120 0.0849 0.160 0.0688 0.501 0.499 Uva_DnaRep.Early_Ovr 1575964 (0.230) 5278776 (0.770) -0.597 0 0.790 0 0.230 0.770 Uva_DnaRep.Mid_Ovr 3980213 (0.514) 3758846 (0.486) -0.098 0.1485 0.129 0.1303 0.514 0.486 Uva_DnaRep.Late_Ovr 7157940 (0.898) 813481 (0.102) 0.576 0 -0.763 0 0.898 0.102 Uva_DnaRep.Pans_Ovr 3334773 (0.636) 1908720 (0.364) 0.116 0.2099 -0.154 0.1865 0.636 0.364 MSA_strict_Ovr 341415 (0.477) 374909 (0.523) -0.164 0.0221 0.217 0.0132 0.477 0.523 MSA_moderate_Ovr 724540 (0.491) 752082 (0.509) -0.139 0.0101 0.184 0.0056 0.491 0.509 MSA_loose_Ovr 1838840 (0.521) 1689132 (0.479) -0.085 0.0127 0.113 0.0076 0.521 0.479 MSA_loose.NonExon_Ovr 1584541 (0.547) 1310414 (0.453) -0.040 0.205 0.052 0.1821 0.547 0.453 MSA_non.coding_Ovr 560518 (0.554) 451139 (0.446) -0.028 0.3682 0.037 0.3526 0.554 0.446 MSA_non.exonic_Ovr 514904 (0.572) 384987 (0.428) 0.004 0.4752 -0.005 0.4726 0.572 0.428
Number of overlapping elements
Gives the number of elements of a given category that overlap each state.Element st0 st1 st0 enr st0 enr p st1 enr st1 enr p ------------------------------ -------------------- -------------------- --------- ---------- ---------- ---------- Num_Gencode_TxStarts 999 (0.353) 1834 (0.647) -0.381 0 0.505 0 Num_Gencode_TxStops 1045 (0.368) 1792 (0.632) -0.354 0 0.468 0 Num_KnownGene_TxStarts 303 (0.371) 513 (0.629) -0.348 0.0001 0.462 0.0002 Num_KnownGene_TxStops 335 (0.408) 487 (0.592) -0.285 0.0012 0.377 0.0009 Num_Ensembl_TxStarts 252 (0.358) 452 (0.642) -0.372 0.0002 0.493 0.0002 Num_Ensembl_TxStops 287 (0.403) 425 (0.597) -0.293 0.0013 0.388 0.001 Num_mRNA_TxStarts 1272 (0.263) 3563 (0.737) -0.538 0 0.713 0 Num_mRNA_TxStops 1318 (0.274) 3486 (0.726) -0.519 0 0.687 0.0001 Num_Spliced.EST_TxStarts 22670 (0.164) 115705 (0.836) -0.713 0.0001 0.944 0.0008 Num_Spliced.EST_TxStops 19325 (0.208) 73374 (0.792) -0.634 0 0.840 0.0001 Num_CpG_Islands 138 (0.271) 371 (0.729) -0.524 0 0.695 0.0001 Num_SNPs 63941 (0.612) 40562 (0.388) 0.074 0.0225 -0.098 0.0129 Num_Nhgri_Chip.GM 507 (0.376) 842 (0.624) -0.340 0 0.451 0 Num_UWDHS.CAP.GM 922 (0.333) 1846 (0.667) -0.415 0 0.550 0 Num_UWDHS.QCP.GM 190 (0.344) 362 (0.656) -0.396 0.0053 0.525 0.0017 Num_UWDHS.QCP.HeLa 228 (0.325) 474 (0.675) -0.430 0.0039 0.570 0.001 Num_UVaDnaRep.OriginsPred 157 (0.588) 110 (0.412) 0.032 0.2484 -0.042 0.2258 Num_ChipCenters.H3K4me1.GM 797 (0.469) 901 (0.531) -0.176 0 0.234 0 Num_ChipCenters.H3K4me1.HeLa 198 (0.186) 864 (0.814) -0.673 0 0.891 0 Num_ChipCenters.H3K4me2.GM 624 (0.442) 789 (0.558) -0.225 0 0.298 0 Num_ChipCenters.H3K4me2.HeLa 97 (0.170) 475 (0.830) -0.702 0 0.931 0 Num_ChipCenters.H3K4me3.GM 442 (0.396) 674 (0.604) -0.305 0 0.404 0 Num_ChipCenters.H3K4me3.HeLa 121 (0.212) 451 (0.788) -0.629 0 0.833 0 Num_ChipCenters.H3ac.GM 330 (0.382) 533 (0.618) -0.329 0 0.436 0 Num_ChipCenters.H3ac.HeLa 148 (0.272) 397 (0.728) -0.523 0 0.694 0 Num_ChipCenters.H4ac.GM 581 (0.470) 654 (0.530) -0.174 0.0036 0.231 0.0012 Num_ChipCenters.H4ac.HeLa 132 (0.201) 524 (0.799) -0.647 0 0.857 0 Num_MSA_strict 14659 (0.479) 15955 (0.521) -0.160 0.0079 0.212 0.0036 Num_MSA_moderate 18871 (0.513) 17883 (0.487) -0.099 0.0116 0.131 0.0066 Num_MSA_loose 53197 (0.531) 47061 (0.469) -0.069 0.0086 0.091 0.0038 Num_MSA_loose.NonExon 51715 (0.523) 47168 (0.477) -0.082 0.0054 0.109 0.0022 Num_MSA_non.coding 17789 (0.525) 16127 (0.475) -0.080 0.0592 0.105 0.0418 Num_MSA_non.exonic 16749 (0.537) 14447 (0.463) -0.058 0.142 0.077 0.1148
Enrichment plots
Plot of enrichment values in each state for various elements.
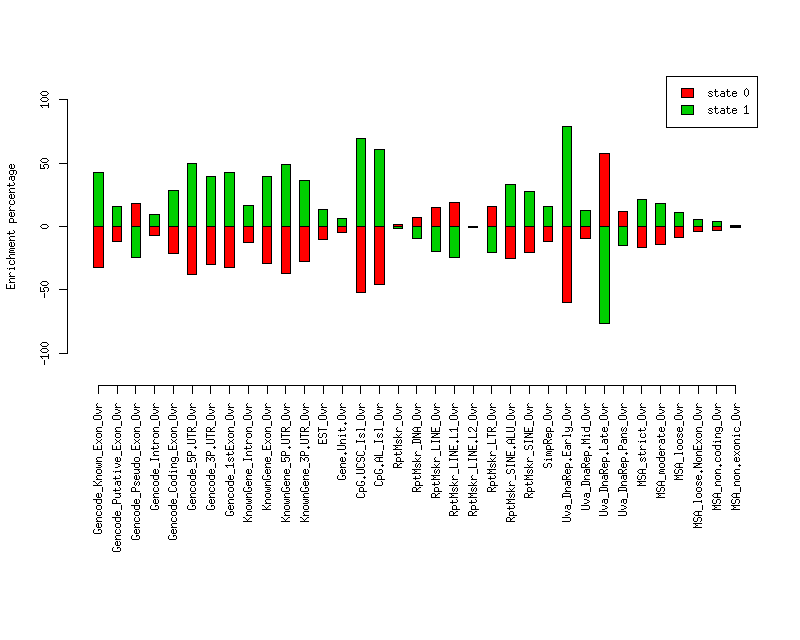
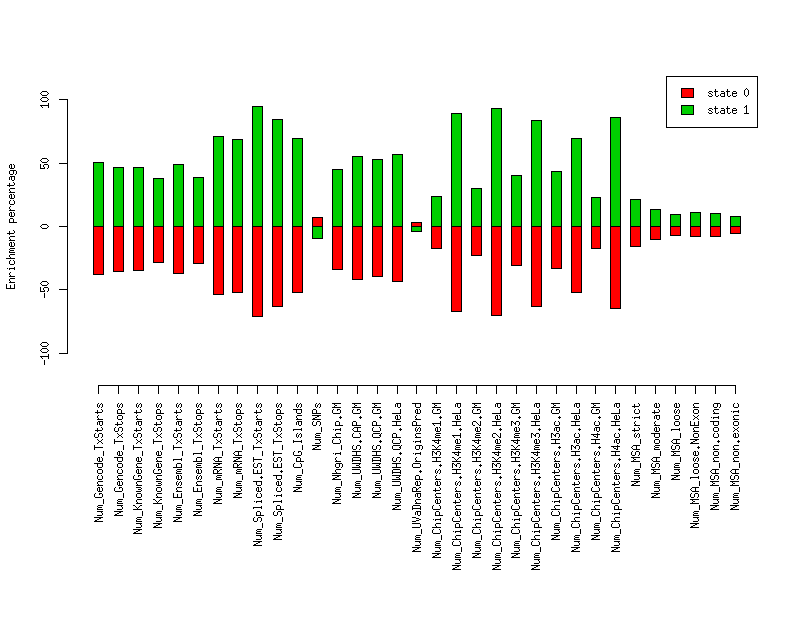
Distances of elements to segment boundaries
Average and median absolute value over all boundaries.
Statistic avg(|d|) med(|d|) ------------------------- ------------ ------------ Gencode_5P_Dist_Known 2103881.19 16540.0 Gencode_3P_Dist_Known 2102473.37 15685.0 Gencode_5P_Dist_Putative 3591176.63 107841.0 Gencode_3P_Dist_Putative 3590171.95 105057.0 Gencode_5P_Dist_Pseudo 986176.94 85375.0 Gencode_3P_Dist_Pseudo 986257.29 85645.5 Gencode_TxStop_Dist 364359.26 10303.0 Gencode_TxStart_Dist 365746.26 9184.0 Gencode_5P.UTR_Dist 2111127.69 14546.0 Gencode_1stExon_Dist 366465.43 9543.0 KnownGene_3P_Dist 105395.33 33359.0 KnownGene_5P_Dist 97221.62 29486.0 Ensemble_5P_Dist 96861.68 29430.0 Refseq_5P_Dist 96394.29 36452.0 mRNA_5P_Dist 53637.49 13275.0 mRNA_3P_Dist 52552.25 18032.0 Spliced.EST_5P_Dist 14733.89 2225.0 Spliced.EST_3P_Dist 12952.46 2170.0 CpG.UCSC_Isl_Dist 99605.08 37407.0 CpG.AL_Isl_Dist 1343861.43 22451.0 Nhgri_Chip.GM_Dist 1314837.39 13145.0 UWDHS.CAP.GM_Dist 423848.08 11452.0 UWDHS.QCP.GM_Dist 18567645.14 78957.0 UWDHS.QCP.HeLa_Dist 18559377.61 54604.0 UVaDnaRep.OriginsPred_Dist 153088.34 40205.0 Sanger_ChipCenters.H3K4me1.GM_Dist 22973.97 10269.0 Sanger_ChipCenters.H3K4me1.HeLa_Dist 382927.43 16411.0 Sanger_ChipCenters.H3K4me2.GM_Dist 28459.46 12041.0 Sanger_ChipCenters.H3K4me2.HeLa_Dist 2640221.00 36933.0 Sanger_ChipCenters.H3K4me3.GM_Dist 39612.20 16200.0 Sanger_ChipCenters.H3K4me3.HeLa_Dist 3119990.67 36059.0 Sanger_ChipCenters.H3ac.GM_Dist 69690.61 25118.0 Sanger_ChipCenters.H3ac.HeLa_Dist 2624150.68 47097.0 Sanger_ChipCenters.H4ac.GM_Dist 58859.87 21860.0 Sanger_ChipCenters.H4ac.HeLa_Dist 1814297.20 42057.0 Uva_DnaRep.Early_Dist 8179885.52 88534.0 Uva_DnaRep.Mid_Dist 4038311.31 46199.0 Uva_DnaRep.Late_Dist 820564.86 87477.0 Uva_DnaRep.PanS_Dist 4130827.79 193926.5 MSA_non.exonic_Dist 6013.31 564.0