Annotation results
H3ac/H3K27me3/TR50 (wavelet) 3-track, 2-state segmentation
|
Gene overlapBed files below contain one line for each gene whose Tx start sits inside the given state. Columns are chr, start, stop, gene ID, fraction of gene sitting inside the segment, strand, and segment ID
|
Enrichment of various elements
Total number of bases or elements overlapping each state. Numbers in parentheses are the fractions of the element in that state. Second set of columns gives enrichment over expected (as a fraction). Enrichment p-values, computed by permuting state labels on the fixed segmentation, are also included, with adjusted p-values using method "BH".Element st0 st1 st0 enr st0 enr p st0 adj p st1 enr st1 enr p st1 adj p ------------------------- -------------------- -------------------- --------- ---------- ---------- --------- ---------- ---------- Num_Gencode_TxStarts 1871 (0.691) 838 (0.309) 0.394 0.0014 0.005564 -0.387 0.0001 0.00085 Num_mRNA_TxStarts 3310 (0.717) 1305 (0.283) 0.447 0.0197 0.0319 -0.439 0.0059 0.01254 Num_Spliced.EST_TxStarts 108245 (0.828) 22476 (0.172) 0.671 0.0785 0.1213 -0.659 0.0119 0.02023 EST_Ovr 10742157 (0.565) 8284782 (0.435) 0.139 0.0017 0.005564 -0.137 0.0026 0.005893 CpG.UCSC_Isl_Ovr 254343 (0.674) 123085 (0.326) 0.360 0.0106 0.01897 -0.354 0.0025 0.005893 RptMskr_Ovr 6526095 (0.494) 6693133 (0.506) -0.004 0.4241 0.4791 0.004 0.4201 0.4791 RptMskr_DNA_Ovr 382608 (0.471) 430155 (0.529) -0.050 0.1533 0.2172 0.049 0.1381 0.2041 RptMskr_LINE.L1_Ovr 1693052 (0.413) 2405066 (0.587) -0.166 0.0022 0.005893 0.163 0.0006 0.0034 RptMskr_LINE.L2_Ovr 479687 (0.480) 519108 (0.520) -0.031 0.2875 0.376 0.030 0.2711 0.3687 RptMskr_LTR_Ovr 790152 (0.384) 1265254 (0.616) -0.224 0 0 0.220 0 0 RptMskr_SINE.ALU_Ovr 2510229 (0.663) 1275867 (0.337) 0.338 0.0006 0.0034 -0.332 0.0001 0.00085 SimpRep_Ovr 332051 (0.481) 358683 (0.519) -0.030 0.4105 0.4791 0.029 0.4044 0.4791 MSA_strict_Ovr 433991 (0.608) 280008 (0.392) 0.227 0.0067 0.01266 -0.223 0.0017 0.005564 MSA_moderate_Ovr 857715 (0.586) 605314 (0.414) 0.183 0.0064 0.01266 -0.180 0.0025 0.005893 MSA_loose_Ovr 1963768 (0.564) 1520991 (0.436) 0.137 0.0018 0.005564 -0.135 0.0013 0.005564 MSA_non.exonic_Ovr 437334 (0.489) 456398 (0.511) -0.012 0.4509 0.4791 0.012 0.4472 0.4791 DiseaseVarMerge_Ovr 1509 (0.497) 1527 (0.503) 0.003 0.4995 0.4995 -0.003 0.4992 0.4995
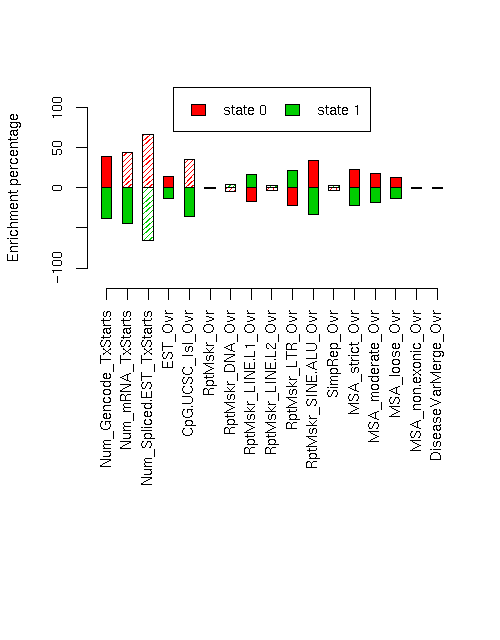
Distances of elements to segment boundaries
Average and median absolute value over all boundaries.
Statistic avg(|d|) med(|d|) ------------------------- ------------ ------------ Gencode_5P_Dist_Known 2108304.45 21254.0 Gencode_3P_Dist_Known 2111493.90 17569.0 Gencode_5P_Dist_Putative 3480968.34 120345.0 Gencode_3P_Dist_Putative 3477983.81 113889.0 Gencode_5P_Dist_Pseudo 2126153.46 90401.5 Gencode_3P_Dist_Pseudo 2125905.88 89192.0 Gencode_TxStop_Dist 523102.66 9903.0 Gencode_TxStart_Dist 523503.15 12311.0 Gencode_5P.UTR_Dist 2122968.08 20886.0 Gencode_1stExon_Dist 524533.71 12649.0 KnownGene_3P_Dist 111130.80 44986.0 KnownGene_5P_Dist 107600.47 38440.0 Ensemble_5P_Dist 107678.10 37076.0 Refseq_5P_Dist 112562.63 46558.0 mRNA_5P_Dist 61144.80 15865.0 mRNA_3P_Dist 59572.46 20621.0 Spliced.EST_5P_Dist 19077.55 3646.0 Spliced.EST_3P_Dist 16886.35 3075.0 CpG.UCSC_Isl_Dist 120808.87 52009.0 CpG.AL_Isl_Dist 1005883.57 26530.0 Nhgri_Chip.GM_Dist 973069.99 17000.0 UWDHS.CAP.GM_Dist 603812.09 15081.0 UWDHS.QCP.GM_Dist 19565438.41 93544.0 UWDHS.QCP.HeLa_Dist 19559723.36 92843.0 UVaDnaRep.OriginsPred_Dist 58210.67 47488.0 Sanger_ChipCenters.H3K4me1.GM_Dist 27162.46 9990.0 Sanger_ChipCenters.H3K4me1.HeLa_Dist 559906.57 16447.0 Sanger_ChipCenters.H3K4me2.GM_Dist 34785.34 12139.0 Sanger_ChipCenters.H3K4me2.HeLa_Dist 2885844.57 45773.0 Sanger_ChipCenters.H3K4me3.GM_Dist 52796.96 22904.0 Sanger_ChipCenters.H3K4me3.HeLa_Dist 4330830.01 40866.0 Sanger_ChipCenters.H3ac.GM_Dist 75250.46 37461.0 Sanger_ChipCenters.H3ac.HeLa_Dist 3439247.53 69455.0 Sanger_ChipCenters.H4ac.GM_Dist 61364.80 25661.0 Sanger_ChipCenters.H4ac.HeLa_Dist 1678093.57 53623.0 Uva_DnaRep.Early_Dist 10858099.80 103774.5 Uva_DnaRep.Mid_Dist 5417048.30 33196.0 Uva_DnaRep.Late_Dist 1129005.12 65124.0 Uva_DnaRep.PanS_Dist 6568766.84 172196.5 MSA_non.exonic_Dist 10243.67 6974.0