High-confidence active/inactive segments
Following are segments whose state assignments agree across all 4 different segmentations.- one-track segmentation of 51.2kb wavelet-smoothed Affy RNA
- one-track segmentation of 64kb wavelet-smoothed Sanger H3ac
- one-track segmentation of 64kb wavelet-smoothed UCSD H3K27me3
- one-track segmentation of TR50
- Number of segments
- Inactive: 43
- Active: 51
- Total size (bp)
- Inactive: 4241811
- Active: 6087597
- Avg. segment length
- Inactive: 98647
- Active: 119365
- Median segment length
- Inactive: 73999
- Active: 72775
- Minimum segment length
- Inactive: 2599
- Active: 1174
- Maximum segment length
- Inactive: 306283
- Active: 495865
Gene overlap
Bed files below contain one line for each gene whose Tx start sits inside the given state. Columns are chr, start, stop, gene ID, fraction of gene sitting inside the segment, strand, and segment ID
- Overlap with active state
- GO term analysis -- biological process
- GO term analysis -- molecular function
- GO term analysis -- cellular components
- Overlap with inactive state
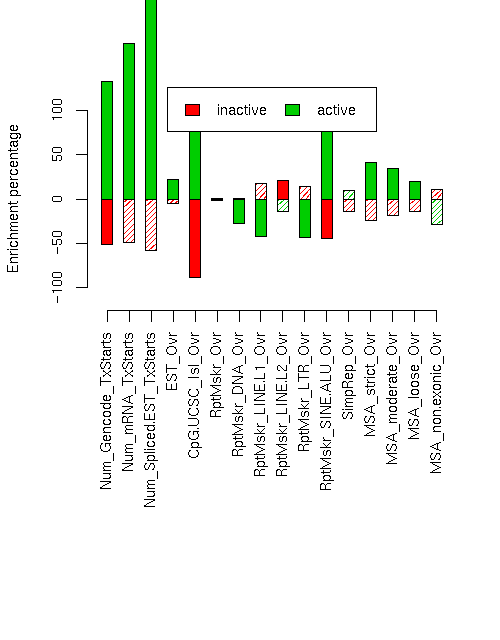
Enrichment/depletion of various elements
Enrichment p-values are computed by permuting state labels on the fixed segmentation 10000 times, with adjusted p-values using method "BH".Element inactive active inact enr inact enr p inact adj p act enr act enr p act adj p ------------------------- ------------ ------------ --------- ----------- ----------- --------- ---------- ---------- Num_Gencode_TxStarts 196 1339 -0.512 0.0006 0.0016 1.325 0 0 Num_mRNA_TxStarts 346 2703 -0.494 0.019 0.032 1.753 0 0 Num_Spliced.EST_TxStarts 8168 96898 -0.583 0.1413 0.1809 2.448 0 0 EST_Ovr 2619036 4838217 -0.053 0.2386 0.2828 0.219 0 0 CpG.UCSC_Isl_Ovr 6709 172629 -0.880 0 0 1.153 0 0 RptMskr_Ovr 1887499 2751800 -0.013 0.3206 0.342 0.003 0.4596 0.4744 RptMskr_DNA_Ovr 116887 121057 0.001 0.4925 0.4925 -0.278 0.0001 0.00032 RptMskr_LINE.L1_Ovr 700111 494945 0.173 0.0183 0.032 -0.422 0 0 RptMskr_LINE.L2_Ovr 172048 176164 0.207 0.0009 0.002057 -0.139 0.0263 0.04208 RptMskr_LTR_Ovr 340543 244932 0.137 0.0293 0.04262 -0.430 0 0 RptMskr_SINE.ALU_Ovr 304553 1436793 -0.440 0.0003 0.0008727 0.841 0 0 SimpRep_Ovr 91268 167620 -0.142 0.211 0.2597 0.098 0.306 0.3377 MSA_strict_Ovr 76840 205982 -0.242 0.0343 0.04772 0.415 0.0026 0.005547 MSA_moderate_Ovr 170597 402256 -0.184 0.0449 0.05987 0.341 0.0009 0.002057 MSA_loose_Ovr 430412 859729 -0.139 0.0281 0.04262 0.198 0.0041 0.0082 MSA_non.exonic_Ovr 141673 130217 0.112 0.2532 0.2894 -0.288 0.0185 0.032