High-confidence active/inactive segments
Following are segments whose state assignments agree across all 6different segmentations.- one-track segmentation of 64kb wavelet-smoothed Sanger H4ac
- one-track segmentation of 64kb wavelet-smoothed UCSD H3K27me3
- one-track segmentation of 64kb wavelet-smoothed MSA non-exonic, 3kb sliding window
- one-track segmentation of TR50
- five-track segmentation of all of the above, plus Affy RNA
- Number of segments
- Inactive: 25
- Active: 50
- Total size (bp)
- Inactive: 2236142
- Active: 4775633
- Avg. segment length
- Inactive: 89446
- Active: 95513
- Median segment length
- Inactive: 57249
- Active: 70387.5
- Minimum segment length
- Inactive: 9817
- Active: 1174
- Maximum segment length
- Inactive: 230649
- Active: 495865
Gene overlap
Bed files below contain one line for each gene whose Tx start sits inside the given state. Columns are chr, start, stop, gene ID, fraction of gene sitting inside the segment, strand, and segment ID
- Overlap with active state
- GO term analysis -- biological process
- GO term analysis -- molecular function
- GO term analysis -- cellular components
- Overlap with inactive state
Enrichment/depletion of various elements
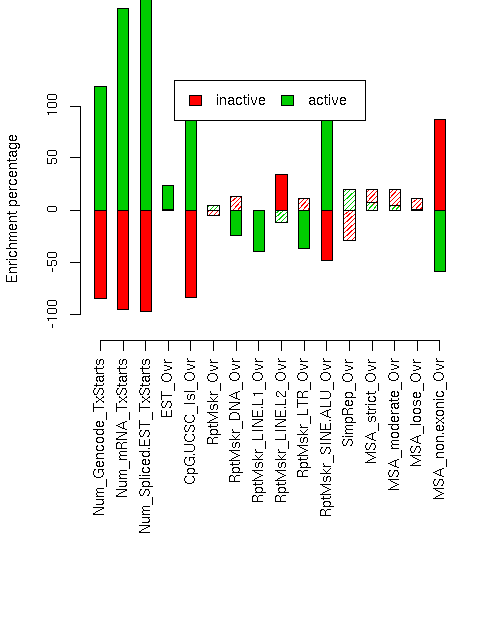
Enrichment p-values are computed by permuting state labels on the fixed segmentation 10000 times, with adjusted p-values using method "BH".Element inactive active inact enr inact enr p inact adj p act enr act enr p act adj p ------------------------- ------------ ------------ --------- ----------- ----------- --------- ---------- ---------- Num_Gencode_TxStarts 32 988 -0.849 0 0 1.187 0 0 Num_mRNA_TxStarts 17 2262 -0.953 0 0 1.936 0 0 Num_Spliced.EST_TxStarts 297 89524 -0.971 0 0 3.061 0 0 EST_Ovr 1468423 3836038 0.007 0.4652 0.4827 0.232 0.0001 0.0002133 CpG.UCSC_Isl_Ovr 4813 124376 -0.837 0 0 0.977 0 0 RptMskr_Ovr 957791 2250056 -0.050 0.0779 0.1187 0.045 0.0526 0.08859 RptMskr_DNA_Ovr 69611 99499 0.131 0.0603 0.09648 -0.243 0 0 RptMskr_LINE.L1_Ovr 311893 406427 -0.009 0.4676 0.4827 -0.395 0 0 RptMskr_LINE.L2_Ovr 101058 140876 0.345 0 0 -0.122 0.0484 0.08604 RptMskr_LTR_Ovr 174554 211012 0.106 0.1087 0.1512 -0.374 0 0 RptMskr_SINE.ALU_Ovr 148407 1177535 -0.482 0.0004 0.0008 0.923 0 0 SimpRep_Ovr 39793 143100 -0.291 0.1351 0.1717 0.194 0.1755 0.208 MSA_strict_Ovr 64084 121928 0.199 0.1395 0.1717 0.068 0.3088 0.3529 MSA_moderate_Ovr 131931 244526 0.197 0.0926 0.1347 0.039 0.352 0.3884 MSA_loose_Ovr 293720 563551 0.114 0.1176 0.1568 0.001 0.4874 0.4874 MSA_non.exonic_Ovr 125614 58388 0.871 0.0005 0.0009412 -0.593 0 0