Annotation results
UCSD Chip data, H3K27me3, HeLa, 64kb wavelet smooth data, 2-state segmentation (state 0 = active!)
|
Gene overlapBed files below contain one line for each gene whose Tx start sits inside the given state. Columns are chr, start, stop, gene ID, fraction of gene sitting inside the segment, strand, and segment ID
|
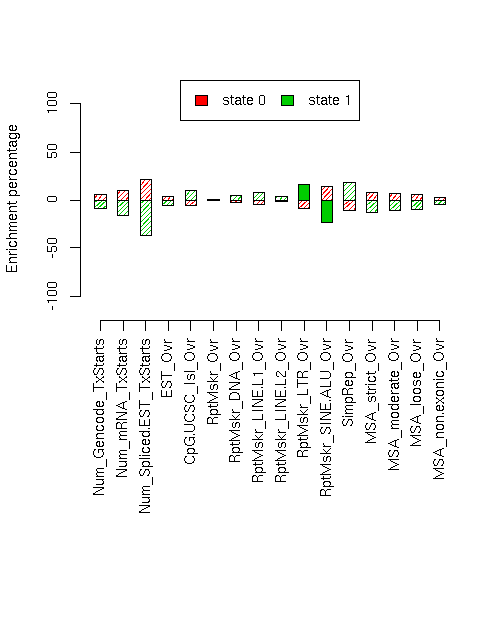
Enrichment of various elements
Total number of bases or elements overlapping each state. Numbers in parentheses are the fractions of the element in that state. Second set of columns gives enrichment over expected (as a fraction). Enrichment p-values, computed by permuting state labels on the fixed segmentation, are also included, with adjusted p-values using method "BH".Element st0 st1 st0 enr st0 enr p st0 adj p st1 enr st1 enr p st1 adj p ------------------------- -------------------- -------------------- --------- ---------- ---------- --------- ---------- ---------- Num_Gencode_TxStarts 1889 (0.666) 946 (0.334) 0.053 0.3129 0.4397 -0.091 0.2318 0.4035 Num_mRNA_TxStarts 3350 (0.693) 1486 (0.307) 0.094 0.4272 0.4882 -0.163 0.3474 0.4447 Num_Spliced.EST_TxStarts 106252 (0.768) 32161 (0.232) 0.213 0.5211 0.5211 -0.367 0.4522 0.499 EST_Ovr 12753321 (0.655) 6717104 (0.345) 0.035 0.2094 0.3942 -0.060 0.1265 0.3488 CpG.UCSC_Isl_Ovr 236210 (0.598) 158645 (0.402) -0.055 0.316 0.4397 0.095 0.2492 0.4035 RptMskr_Ovr 8487845 (0.633) 4930580 (0.367) -0.001 0.4875 0.5032 0.001 0.4814 0.5032 RptMskr_DNA_Ovr 506652 (0.616) 316189 (0.384) -0.027 0.2648 0.4035 0.047 0.1709 0.3765 RptMskr_LINE.L1_Ovr 2515460 (0.603) 1654275 (0.397) -0.047 0.1765 0.3765 0.081 0.0882 0.3488 RptMskr_LINE.L2_Ovr 624311 (0.621) 381479 (0.379) -0.019 0.3318 0.4424 0.033 0.2535 0.4035 RptMskr_LTR_Ovr 1204560 (0.575) 888942 (0.425) -0.091 0.0235 0.2376 0.157 0.0035 0.0928 RptMskr_SINE.ALU_Ovr 2753899 (0.720) 1072386 (0.280) 0.137 0.0478 0.3059 -0.236 0.0058 0.0928 SimpRep_Ovr 425392 (0.567) 324902 (0.433) -0.104 0.1308 0.3488 0.180 0.0599 0.3195 MSA_strict_Ovr 487143 (0.680) 229583 (0.320) 0.074 0.1985 0.3942 -0.127 0.1101 0.3488 MSA_moderate_Ovr 995921 (0.674) 481694 (0.326) 0.065 0.1696 0.3765 -0.112 0.0795 0.3488 MSA_loose_Ovr 2363584 (0.669) 1167535 (0.331) 0.058 0.0984 0.3488 -0.099 0.0297 0.2376 MSA_non.exonic_Ovr 585635 (0.651) 314512 (0.349) 0.028 0.4168 0.4882 -0.048 0.3752 0.4618
Distances of elements to segment boundaries
Average and median absolute value over all boundaries.
Statistic avg(|d|) med(|d|) ------------------------- ------------ ------------ Gencode_5P_Dist_Known 2193847.73 18111.0 Gencode_3P_Dist_Known 2193299.37 13505.0 Gencode_5P_Dist_Putative 3594748.04 109883.0 Gencode_3P_Dist_Putative 3593198.26 105084.0 Gencode_5P_Dist_Pseudo 1497712.19 91153.0 Gencode_3P_Dist_Pseudo 1497532.63 91679.0 Gencode_TxStop_Dist 709834.90 7566.0 Gencode_TxStart_Dist 712393.16 8503.0 Gencode_5P.UTR_Dist 2209526.81 17996.0 Gencode_1stExon_Dist 713330.50 9109.0 KnownGene_3P_Dist 103287.55 33326.0 KnownGene_5P_Dist 99666.19 31195.0 Ensemble_5P_Dist 99652.51 29769.0 Refseq_5P_Dist 104971.19 41552.0 mRNA_5P_Dist 57949.16 14214.0 mRNA_3P_Dist 54734.16 17112.0 Spliced.EST_5P_Dist 19647.92 4200.0 Spliced.EST_3P_Dist 16357.48 2731.0 CpG.UCSC_Isl_Dist 121213.92 48149.0 CpG.AL_Isl_Dist 945638.61 21188.0 Nhgri_Chip.GM_Dist 905049.95 13636.0 UWDHS.CAP.GM_Dist 568795.80 16056.0 UWDHS.QCP.GM_Dist 21166098.82 93704.0 UWDHS.QCP.HeLa_Dist 21158805.30 85196.0 UVaDnaRep.OriginsPred_Dist 178666.07 41023.0 Sanger_ChipCenters.H3K4me1.GM_Dist 26982.22 10785.0 Sanger_ChipCenters.H3K4me1.HeLa_Dist 744807.54 14196.0 Sanger_ChipCenters.H3K4me2.GM_Dist 35214.76 12973.0 Sanger_ChipCenters.H3K4me2.HeLa_Dist 3101991.00 38553.0 Sanger_ChipCenters.H3K4me3.GM_Dist 47537.49 17632.0 Sanger_ChipCenters.H3K4me3.HeLa_Dist 3756224.31 32436.0 Sanger_ChipCenters.H3ac.GM_Dist 78071.71 30885.0 Sanger_ChipCenters.H3ac.HeLa_Dist 2510037.72 58002.0 Sanger_ChipCenters.H4ac.GM_Dist 64845.82 26826.0 Sanger_ChipCenters.H4ac.HeLa_Dist 1569454.23 51666.0 Uva_DnaRep.Early_Dist 11101354.53 142697.0 Uva_DnaRep.Mid_Dist 5537731.89 56930.0 Uva_DnaRep.Late_Dist 1076867.67 68135.0 Uva_DnaRep.PanS_Dist 5357751.44 151935.0 MSA_non.exonic_Dist 9380.50 6908.5