Annotation results
SangerChip data, H4ac, HeLa, 64kb wavelet smooth data, 2-state segmentation
|
Gene overlapBed files below contain one line for each gene whose Tx start sits inside the given state. Columns are chr, start, stop, gene ID, fraction of gene sitting inside the segment, strand, and segment ID
|
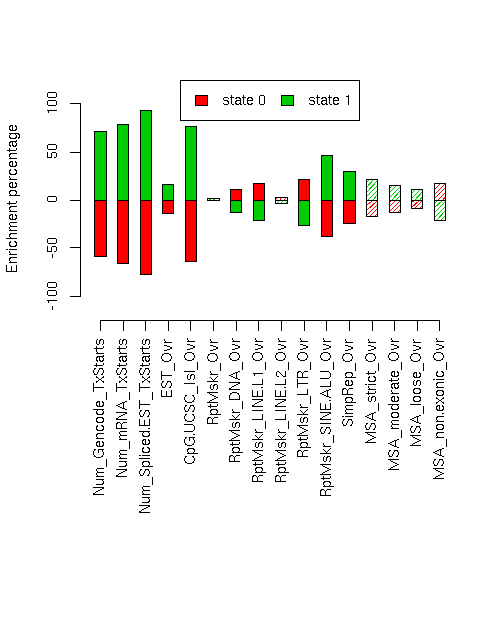
Enrichment of various elements
Total number of bases or elements overlapping each state. Numbers in parentheses are the fractions of the element in that state. Second set of columns gives enrichment over expected (as a fraction). Enrichment p-values, computed by permuting state labels on the fixed segmentation, are also included, with adjusted p-values using method "BH".Element st0 st1 st0 enr st0 enr p st0 adj p st1 enr st1 enr p st1 adj p ------------------------- -------------------- -------------------- --------- ---------- ---------- --------- ---------- ---------- Num_Gencode_TxStarts 631 (0.223) 2202 (0.777) -0.592 0 0 0.709 0 0 Num_mRNA_TxStarts 897 (0.186) 3938 (0.814) -0.660 0 0 0.791 0 0 Num_Spliced.EST_TxStarts 16819 (0.122) 121557 (0.878) -0.777 0 0 0.932 0.0004 0.0008533 EST_Ovr 9166128 (0.471) 10297934 (0.529) -0.136 0.0007 0.0014 0.163 0.0004 0.0008533 CpG.UCSC_Isl_Ovr 77550 (0.196) 317429 (0.804) -0.640 0 0 0.767 0 0 RptMskr_Ovr 7251514 (0.540) 6167699 (0.460) -0.009 0.31 0.31 0.011 0.2932 0.3027 RptMskr_DNA_Ovr 497692 (0.605) 324706 (0.395) 0.110 0.0064 0.01024 -0.132 0.0036 0.0064 RptMskr_LINE.L1_Ovr 2676836 (0.642) 1494313 (0.358) 0.177 0.0004 0.0008533 -0.212 0.0001 0.0002667 RptMskr_LINE.L2_Ovr 562403 (0.560) 442695 (0.440) 0.026 0.2903 0.3027 -0.031 0.2779 0.3027 RptMskr_LTR_Ovr 1391682 (0.665) 702644 (0.335) 0.219 0 0 -0.262 0 0 RptMskr_SINE.ALU_Ovr 1284105 (0.336) 2542215 (0.664) -0.385 0 0 0.461 0 0 SimpRep_Ovr 308453 (0.411) 442476 (0.589) -0.247 0.0036 0.0064 0.296 0.0048 0.008084 MSA_strict_Ovr 322706 (0.450) 393623 (0.550) -0.174 0.0234 0.0288 0.208 0.0182 0.02532 MSA_moderate_Ovr 700630 (0.474) 776003 (0.526) -0.130 0.0262 0.03105 0.156 0.0199 0.02653 MSA_loose_Ovr 1747124 (0.495) 1780859 (0.505) -0.092 0.0148 0.02153 0.110 0.0112 0.01707 MSA_non.exonic_Ovr 576270 (0.640) 323625 (0.360) 0.174 0.0355 0.04057 -0.209 0.0219 0.02803
Distances of elements to segment boundaries
Average and median absolute value over all boundaries.
Statistic avg(|d|) med(|d|) ------------------------- ------------ ------------ Gencode_5P_Dist_Known 1883228.16 21831.5 Gencode_3P_Dist_Known 1886013.74 20302.0 Gencode_5P_Dist_Putative 3573066.85 119160.0 Gencode_3P_Dist_Putative 3572028.51 113846.0 Gencode_5P_Dist_Pseudo 1464746.13 89391.0 Gencode_3P_Dist_Pseudo 1464300.39 89559.0 Gencode_TxStop_Dist 467008.81 13620.5 Gencode_TxStart_Dist 469003.28 14466.5 Gencode_5P.UTR_Dist 1895998.62 20632.0 Gencode_1stExon_Dist 469888.92 14690.5 KnownGene_3P_Dist 95961.66 35383.5 KnownGene_5P_Dist 91981.53 30811.5 Ensemble_5P_Dist 91180.56 30538.0 Refseq_5P_Dist 99281.63 40438.0 mRNA_5P_Dist 53365.13 14371.5 mRNA_3P_Dist 52167.74 21032.0 Spliced.EST_5P_Dist 17785.17 4382.5 Spliced.EST_3P_Dist 15438.77 3429.0 CpG.UCSC_Isl_Dist 103557.88 37561.0 CpG.AL_Isl_Dist 898207.51 23379.5 Nhgri_Chip.GM_Dist 868014.70 12672.5 UWDHS.CAP.GM_Dist 542744.42 14619.5 UWDHS.QCP.GM_Dist 19432272.26 95435.0 UWDHS.QCP.HeLa_Dist 19425543.76 70639.5 UVaDnaRep.OriginsPred_Dist 130620.97 42320.5 Sanger_ChipCenters.H3K4me1.GM_Dist 27210.01 9418.5 Sanger_ChipCenters.H3K4me1.HeLa_Dist 492642.72 19149.5 Sanger_ChipCenters.H3K4me2.GM_Dist 33665.36 11641.5 Sanger_ChipCenters.H3K4me2.HeLa_Dist 2576216.73 40758.5 Sanger_ChipCenters.H3K4me3.GM_Dist 45577.31 16121.0 Sanger_ChipCenters.H3K4me3.HeLa_Dist 3421519.31 36910.5 Sanger_ChipCenters.H3ac.GM_Dist 79352.23 29398.0 Sanger_ChipCenters.H3ac.HeLa_Dist 2477835.05 50289.0 Sanger_ChipCenters.H4ac.GM_Dist 66446.61 25656.0 Sanger_ChipCenters.H4ac.HeLa_Dist 1498605.73 45795.0 Uva_DnaRep.Early_Dist 9315695.24 120328.0 Uva_DnaRep.Mid_Dist 5191078.39 55501.0 Uva_DnaRep.Late_Dist 552794.68 70411.5 Uva_DnaRep.PanS_Dist 5129837.44 173276.0 MSA_non.exonic_Dist 6877.00 998.0