Annotation results
SangerChip data, H3ac, HeLa, 64kb wavelet smooth data, 2-state segmentation
|
Gene overlapBed files below contain one line for each gene whose Tx start sits inside the given state. Columns are chr, start, stop, gene ID, fraction of gene sitting inside the segment, strand, and segment ID
|
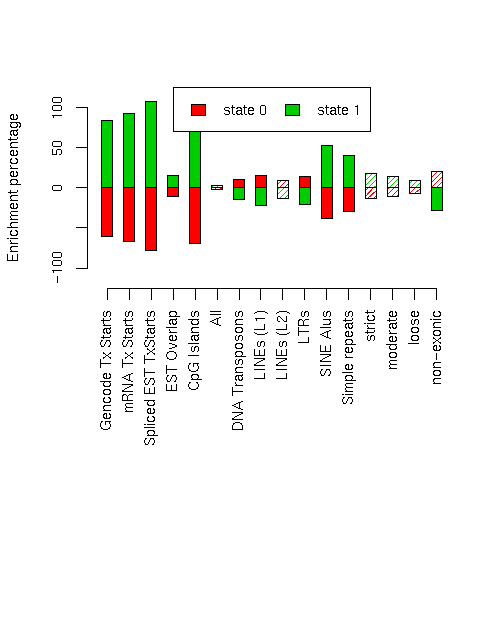
Enrichment of various elements
Total number of bases or elements overlapping each state. Numbers in parentheses are the fractions of the element in that state. Second set of columns gives enrichment over expected (as a fraction). Enrichment p-values, computed by permuting state labels on the fixed segmentation, are also included, with adjusted p-values using method "BH".Element st0 st1 st0 enr st0 enr p st0 adj p st1 enr st1 enr p st1 adj p ------------------------- -------------------- -------------------- --------- ---------- ---------- --------- ---------- ---------- Num_Gencode_TxStarts 645 (0.228) 2188 (0.772) -0.608 0 0 0.843 0 0 Num_mRNA_TxStarts 924 (0.191) 3911 (0.809) -0.671 0 0 0.931 0 0 Num_Spliced.EST_TxStarts 17604 (0.127) 120771 (0.873) -0.781 0 0 1.083 0.0001 0.0002909 EST_Ovr 10061831 (0.517) 9402217 (0.483) -0.110 0.0038 0.0064 0.153 0.0006 0.001477 CpG.UCSC_Isl_Ovr 71197 (0.180) 323782 (0.820) -0.690 0 0 0.957 0 0 RptMskr_Ovr 7654161 (0.570) 5765053 (0.430) -0.018 0.1422 0.1422 0.025 0.1166 0.1204 RptMskr_DNA_Ovr 526093 (0.640) 296305 (0.360) 0.101 0.0063 0.0096 -0.140 0.0024 0.004267 RptMskr_LINE.L1_Ovr 2803611 (0.672) 1367542 (0.328) 0.157 0.0009 0.002057 -0.217 0.0004 0.001067 RptMskr_LINE.L2_Ovr 637733 (0.634) 367365 (0.366) 0.092 0.0215 0.02867 -0.128 0.0138 0.0192 RptMskr_LTR_Ovr 1394577 (0.666) 699746 (0.334) 0.146 0.001 0.002133 -0.203 0 0 RptMskr_SINE.ALU_Ovr 1374171 (0.359) 2452149 (0.641) -0.382 0 0 0.530 0 0 SimpRep_Ovr 307972 (0.410) 442957 (0.590) -0.294 0.0013 0.002447 0.408 0.0013 0.002447 MSA_strict_Ovr 360216 (0.503) 356115 (0.497) -0.135 0.0661 0.07051 0.187 0.0382 0.04527 MSA_moderate_Ovr 767268 (0.520) 709365 (0.480) -0.106 0.0638 0.0704 0.147 0.0375 0.04527 MSA_loose_Ovr 1905571 (0.540) 1622414 (0.460) -0.070 0.0533 0.06091 0.098 0.0286 0.03661 MSA_non.exonic_Ovr 630174 (0.700) 269720 (0.300) 0.205 0.0122 0.01775 -0.285 0.0047 0.00752
Distances of elements to segment boundaries
Average and median absolute value over all boundaries.
Statistic avg(|d|) med(|d|) ------------------------- ------------ ------------ Gencode_5P_Dist_Known 2557230.28 21439.5 Gencode_3P_Dist_Known 2560447.88 22187.5 Gencode_5P_Dist_Putative 4464656.36 104988.0 Gencode_3P_Dist_Putative 4463219.09 107559.5 Gencode_5P_Dist_Pseudo 1390835.83 95436.0 Gencode_3P_Dist_Pseudo 1390609.51 95801.0 Gencode_TxStop_Dist 441613.16 12636.0 Gencode_TxStart_Dist 442721.29 13816.0 Gencode_5P.UTR_Dist 2568916.02 20632.0 Gencode_1stExon_Dist 443558.81 14756.0 KnownGene_3P_Dist 117341.49 35887.0 KnownGene_5P_Dist 108840.14 37063.0 Ensemble_5P_Dist 107637.83 36076.0 Refseq_5P_Dist 104569.73 42503.5 mRNA_5P_Dist 60034.51 16307.0 mRNA_3P_Dist 57813.65 20684.5 Spliced.EST_5P_Dist 17356.55 4289.0 Spliced.EST_3P_Dist 15761.02 3515.5 CpG.UCSC_Isl_Dist 112152.55 45614.5 CpG.AL_Isl_Dist 1629685.31 27134.0 Nhgri_Chip.GM_Dist 1602433.46 16015.5 UWDHS.CAP.GM_Dist 512835.31 19607.5 UWDHS.QCP.GM_Dist 22087952.22 86250.5 UWDHS.QCP.HeLa_Dist 22079863.37 60065.0 UVaDnaRep.OriginsPred_Dist 125082.62 41671.0 Sanger_ChipCenters.H3K4me1.GM_Dist 24892.74 9343.5 Sanger_ChipCenters.H3K4me1.HeLa_Dist 472887.23 16484.0 Sanger_ChipCenters.H3K4me2.GM_Dist 32649.84 12655.0 Sanger_ChipCenters.H3K4me2.HeLa_Dist 3220164.16 45724.5 Sanger_ChipCenters.H3K4me3.GM_Dist 47053.32 21073.5 Sanger_ChipCenters.H3K4me3.HeLa_Dist 4011606.46 42801.0 Sanger_ChipCenters.H3ac.GM_Dist 70713.96 37032.5 Sanger_ChipCenters.H3ac.HeLa_Dist 3121863.40 46491.0 Sanger_ChipCenters.H4ac.GM_Dist 60910.96 29215.5 Sanger_ChipCenters.H4ac.HeLa_Dist 2206643.03 47000.5 Uva_DnaRep.Early_Dist 10905695.75 106836.0 Uva_DnaRep.Mid_Dist 4621761.80 43447.5 Uva_DnaRep.Late_Dist 574775.77 71415.5 Uva_DnaRep.PanS_Dist 5848003.40 162881.5 MSA_non.exonic_Dist 10628.11 1452.0