Annotation results
MSA non-exonic, 3kb sliding window, 64kb wavelet smooth data, 2-state segmentation
|
Gene overlapBed files below contain one line for each gene whose Tx start sits inside the given state. Columns are chr, start, stop, gene ID, fraction of gene sitting inside the segment, strand, and segment ID
|
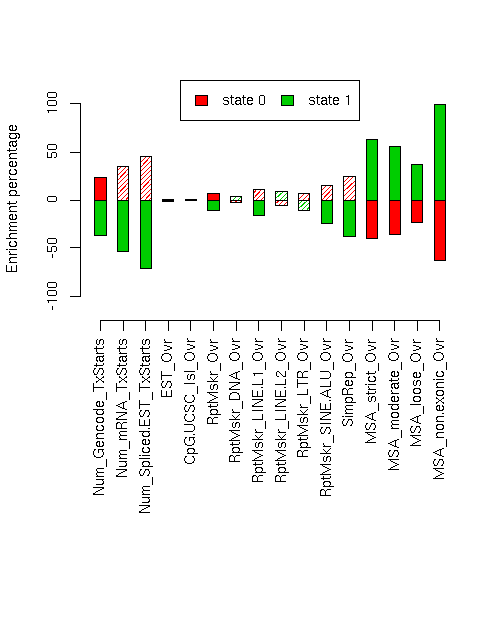
Enrichment of various elements
Total number of bases or elements overlapping each state. Numbers in parentheses are the fractions of the element in that state. Second set of columns gives enrichment over expected (as a fraction). Enrichment p-values, computed by permuting state labels on the fixed segmentation, are also included, with adjusted p-values using method "BH".Element st0 st1 st0 enr st0 enr p st0 adj p st1 enr st1 enr p st1 adj p ------------------------- -------------------- -------------------- --------- ---------- ---------- --------- ---------- ---------- Num_Gencode_TxStarts 2141 (0.754) 697 (0.246) 0.235 0.0082 0.01544 -0.369 0.0015 0.004 Num_mRNA_TxStarts 3972 (0.821) 867 (0.179) 0.344 0.0165 0.0288 -0.540 0.001 0.002909 Num_Spliced.EST_TxStarts 122986 (0.888) 15480 (0.112) 0.455 0.1022 0.1258 -0.713 0.0021 0.005169 EST_Ovr 12042688 (0.616) 7509845 (0.384) 0.009 0.4163 0.4441 -0.014 0.3938 0.4345 CpG.UCSC_Isl_Ovr 242005 (0.612) 153157 (0.388) 0.003 0.5034 0.5034 -0.005 0.5013 0.5034 RptMskr_Ovr 8838355 (0.654) 4685066 (0.346) 0.070 0 0 -0.110 0 0 RptMskr_DNA_Ovr 491104 (0.595) 334638 (0.405) -0.026 0.2616 0.299 0.041 0.2162 0.2562 RptMskr_LINE.L1_Ovr 2847306 (0.674) 1374555 (0.326) 0.104 0.0209 0.03344 -0.164 0.0045 0.0096 RptMskr_LINE.L2_Ovr 580660 (0.576) 427098 (0.424) -0.056 0.0915 0.1171 0.088 0.0484 0.06734 RptMskr_LTR_Ovr 1379636 (0.652) 737848 (0.348) 0.067 0.0614 0.08187 -0.105 0.0274 0.03985 RptMskr_SINE.ALU_Ovr 2707925 (0.704) 1138380 (0.296) 0.153 0.0171 0.0288 -0.240 0.0053 0.0106 SimpRep_Ovr 571088 (0.759) 181502 (0.241) 0.243 0.0227 0.03459 -0.381 0.0035 0.008 MSA_strict_Ovr 261481 (0.365) 455640 (0.635) -0.403 0 0 0.632 0 0 MSA_moderate_Ovr 579713 (0.392) 898834 (0.608) -0.358 0 0 0.561 0 0 MSA_loose_Ovr 1650753 (0.467) 1885248 (0.533) -0.235 0 0 0.369 0 0 MSA_non.exonic_Ovr 200822 (0.223) 699838 (0.777) -0.635 0 0 0.996 0 0
Distances of elements to segment boundaries
Average and median absolute value over all boundaries.
Statistic avg(|d|) med(|d|) ------------------------- ------------ ------------ Gencode_5P_Dist_Known 2159070.23 19589.5 Gencode_3P_Dist_Known 2159943.48 19048.0 Gencode_5P_Dist_Putative 4198795.75 124916.0 Gencode_3P_Dist_Putative 4196868.80 120627.0 Gencode_5P_Dist_Pseudo 1986462.74 121319.0 Gencode_3P_Dist_Pseudo 1986822.39 120063.5 Gencode_TxStop_Dist 842334.69 11260.5 Gencode_TxStart_Dist 843081.87 13284.0 Gencode_5P.UTR_Dist 2185781.95 22385.0 Gencode_1stExon_Dist 844114.00 14233.0 KnownGene_3P_Dist 110242.37 46398.0 KnownGene_5P_Dist 109699.67 40163.5 Ensemble_5P_Dist 116013.51 40610.0 Refseq_5P_Dist 124729.00 48029.0 mRNA_5P_Dist 68198.49 18958.5 mRNA_3P_Dist 62143.83 20485.5 Spliced.EST_5P_Dist 18105.84 6189.5 Spliced.EST_3P_Dist 17397.08 4627.0 CpG.UCSC_Isl_Dist 129895.76 45480.5 CpG.AL_Isl_Dist 854156.90 30025.5 Nhgri_Chip.GM_Dist 817229.90 19234.0 UWDHS.CAP.GM_Dist 811709.63 18747.0 UWDHS.QCP.GM_Dist 22318053.23 9295562.0 UWDHS.QCP.HeLa_Dist 22306996.50 9299898.0 UVaDnaRep.OriginsPred_Dist 88622.56 39164.0 Sanger_ChipCenters.H3K4me1.GM_Dist 24554.33 8513.5 Sanger_ChipCenters.H3K4me1.HeLa_Dist 876823.12 23105.5 Sanger_ChipCenters.H3K4me2.GM_Dist 35113.01 14573.0 Sanger_ChipCenters.H3K4me2.HeLa_Dist 3459782.39 48873.0 Sanger_ChipCenters.H3K4me3.GM_Dist 50555.86 22047.5 Sanger_ChipCenters.H3K4me3.HeLa_Dist 4467545.76 42008.0 Sanger_ChipCenters.H3ac.GM_Dist 79100.35 30761.5 Sanger_ChipCenters.H3ac.HeLa_Dist 2829321.00 60100.5 Sanger_ChipCenters.H4ac.GM_Dist 63596.12 27589.5 Sanger_ChipCenters.H4ac.HeLa_Dist 1419487.88 49845.0 Uva_DnaRep.Early_Dist 8367606.30 146867.0 Uva_DnaRep.Mid_Dist 4448403.96 79087.0 Uva_DnaRep.Late_Dist 388343.50 61577.0 Uva_DnaRep.PanS_Dist 4797395.70 167171.0 MSA_non.exonic_Dist 9521.50 3277.0