Annotation results
Affy RNA Signal data (thresholded), HeLa, 51.2kb wavelet smooth data, 2-state segmentation
|
Gene overlapBed files below contain one line for each gene whose Tx start sits inside the given state. Columns are chr, start, stop, gene ID, fraction of gene sitting inside the segment, strand, and segment ID
|
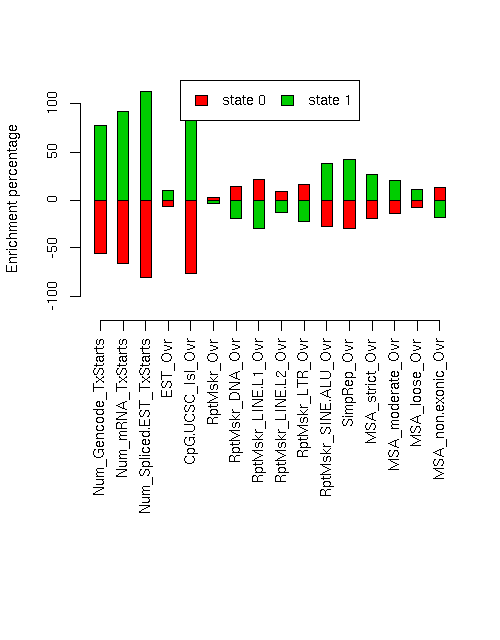
Enrichment of various elements
Total number of bases or elements overlapping each state. Numbers in parentheses are the fractions of the element in that state. Second set of columns gives enrichment over expected (as a fraction). Enrichment p-values, computed by permuting state labels on the fixed segmentation, are also included, with adjusted p-values using method "BH".Element st0 st1 st0 enr st0 enr p st0 adj p st1 enr st1 enr p st1 adj p ------------------------- -------------------- -------------------- --------- ---------- ---------- --------- ---------- ---------- Num_Gencode_TxStarts 712 (0.257) 2056 (0.743) -0.558 0 0 0.776 0 0 Num_mRNA_TxStarts 935 (0.196) 3837 (0.804) -0.663 0 0 0.923 0 0 Num_Spliced.EST_TxStarts 15048 (0.110) 121545 (0.890) -0.811 0 0 1.128 0 0 EST_Ovr 10340852 (0.542) 8745448 (0.458) -0.069 0.0546 0.06016 0.096 0.0277 0.03283 CpG.UCSC_Isl_Ovr 52486 (0.138) 328400 (0.862) -0.763 0 0 1.062 0 0 RptMskr_Ovr 7872674 (0.598) 5299092 (0.402) 0.027 0.0564 0.06016 -0.038 0.0139 0.01934 RptMskr_DNA_Ovr 539287 (0.665) 272264 (0.335) 0.142 0.0003 0.0005333 -0.198 0 0 RptMskr_LINE.L1_Ovr 2894357 (0.706) 1207676 (0.294) 0.213 0 0 -0.296 0 0 RptMskr_LINE.L2_Ovr 624596 (0.635) 358630 (0.365) 0.092 0.0139 0.01934 -0.128 0.0036 0.005486 RptMskr_LTR_Ovr 1383878 (0.674) 669701 (0.326) 0.158 0.0001 0.0002133 -0.220 0 0 RptMskr_SINE.ALU_Ovr 1591902 (0.424) 2162971 (0.576) -0.271 0 0 0.378 0 0 SimpRep_Ovr 291226 (0.407) 424379 (0.593) -0.301 0.0002 0.0004 0.418 0.0003 0.0005333 MSA_strict_Ovr 337462 (0.473) 376402 (0.527) -0.188 0.018 0.02304 0.261 0.0022 0.003705 MSA_moderate_Ovr 730644 (0.498) 735704 (0.502) -0.144 0.0236 0.02905 0.200 0.0029 0.00464 MSA_loose_Ovr 1876137 (0.538) 1613977 (0.462) -0.076 0.0519 0.05931 0.106 0.015 0.02 MSA_non.exonic_Ovr 589805 (0.657) 307721 (0.343) 0.129 0.1029 0.1029 -0.180 0.0666 0.06875
Distances of elements to segment boundaries
Average and median absolute value over all boundaries.
Statistic avg(|d|) med(|d|) ------------------------- ------------ ------------ Gencode_5P_Dist_Known 1348570.44 18125.0 Gencode_3P_Dist_Known 1351579.94 20112.0 Gencode_5P_Dist_Putative 2324944.46 99876.0 Gencode_3P_Dist_Putative 2324490.84 98872.0 Gencode_5P_Dist_Pseudo 1149061.18 75032.0 Gencode_3P_Dist_Pseudo 1149476.75 76305.0 Gencode_TxStop_Dist 336627.68 11576.0 Gencode_TxStart_Dist 335803.44 9161.0 Gencode_5P.UTR_Dist 1357826.01 17889.0 Gencode_1stExon_Dist 336449.64 9352.0 KnownGene_3P_Dist 84290.92 37814.0 KnownGene_5P_Dist 77367.80 27829.0 Ensemble_5P_Dist 74938.50 26841.0 Refseq_5P_Dist 85173.57 33675.0 mRNA_5P_Dist 46562.21 14429.0 mRNA_3P_Dist 46215.15 20554.0 Spliced.EST_5P_Dist 14005.59 4025.0 Spliced.EST_3P_Dist 13068.25 3595.0 CpG.UCSC_Isl_Dist 94174.97 39202.0 CpG.AL_Isl_Dist 645438.69 23425.0 Nhgri_Chip.GM_Dist 622533.40 14813.0 UWDHS.CAP.GM_Dist 878336.19 15630.0 UWDHS.QCP.GM_Dist 19535329.79 34334.0 UWDHS.QCP.HeLa_Dist 19528822.41 43505.0 UVaDnaRep.OriginsPred_Dist 134384.20 42150.0 Sanger_ChipCenters.H3K4me1.GM_Dist 23502.02 9656.0 Sanger_ChipCenters.H3K4me1.HeLa_Dist 360295.00 21518.0 Sanger_ChipCenters.H3K4me2.GM_Dist 28962.46 11361.0 Sanger_ChipCenters.H3K4me2.HeLa_Dist 2067329.96 34517.0 Sanger_ChipCenters.H3K4me3.GM_Dist 42387.86 22953.0 Sanger_ChipCenters.H3K4me3.HeLa_Dist 2762342.64 32167.0 Sanger_ChipCenters.H3ac.GM_Dist 67676.98 28259.0 Sanger_ChipCenters.H3ac.HeLa_Dist 1617264.48 58239.0 Sanger_ChipCenters.H4ac.GM_Dist 61560.36 27936.0 Sanger_ChipCenters.H4ac.HeLa_Dist 1086003.98 49472.0 Uva_DnaRep.Early_Dist 8035580.73 116040.0 Uva_DnaRep.Mid_Dist 3840869.73 63119.0 Uva_DnaRep.Late_Dist 541316.89 66537.0 Uva_DnaRep.PanS_Dist 3828120.57 131733.0 MSA_non.exonic_Dist 6918.18 1815.0