Noble Research Lab

Department of Genome Sciences
University of Washington
Our research group develops and applies computational techniques for
modeling and understanding biological processes at the molecular
level. Our research emphasizes the application of statistical and
machine learning techniques, such as hidden Markov models and support
vector machines. We apply these techniques to various types of
biological data, including protein and DNA sequences, data from
high-throughput genomic assays such as ChIP-seq and Hi-C, and tandem
mass spectrometry. We are currently developing methods for analyzing
shotgun proteomics data, for characterizing protein function,
structure and interactions, and for understanding the structure and
regulatory influence of chromatin.
Inclusion statement

Left to right: Bo Wen, Yuanyuan Zhang, Xiao Wang, Gang Li, Anupama
Jha, Ran Zhang, Borislav Hristov, Gwen Straub, Abel Tesfaye, Bill
Noble
Click here for older pictures.
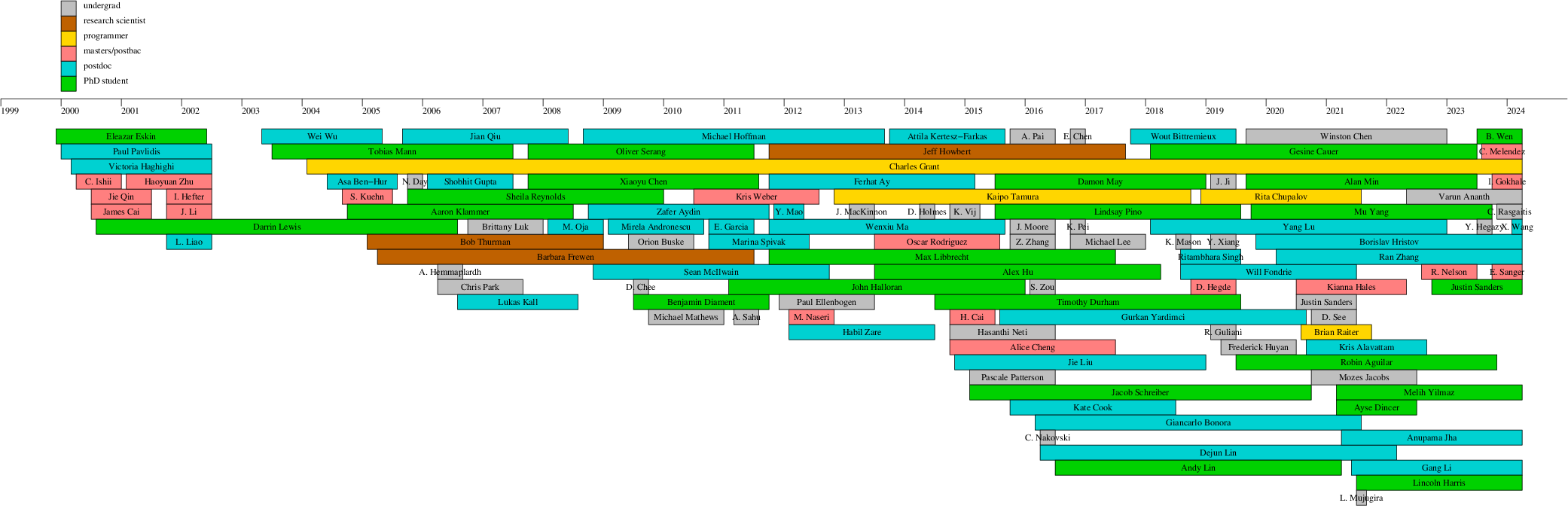
Lab members
Brief bios of current and former lab
members

-
William Stafford
Noble, Professor, Department of Genome Sciences

-
Anupama Jha,
Postdoctoral fellow, Department of Genome Sciences

-
Ghulam Murtaza,
Postdoctoral fellow, Department of Genome Sciences

-
Shengqi Hang, Ph.D. student, Paul G. Allen School of Computer
Science and Engineering

-
Lincoln Harris, Ph.D. student, Department of Genome Sciences

-
Justin Sanders, Ph.D. student, Paul G. Allen School of Computer
Science and Engineering

-
Bo Wen, Ph.D. student, Department of Genome Sciences

-
Annie Staker, Masters student, Masters in Data Sciences

-
Lingwen Xu, Masters student, Paul G. Allen School of Computer
Science and Engineering

-
Xumeng Zhang, Masters student, Statistics

-
Eugene Lin, Research Scientist, Department of Genome Sciences

-
Charles Grant, Senior programmer, Department of Genome Sciences

-
Gwenneth Straub, Bioinformatics Research Scientist, Department
of Genome Sciences

-
Priank Dasgupta, Undergraduate, Engineering

-
Devanshi Desai, Undergraduate, Paul G. Allen School of Computer
Science and Engineering

-
Shanti Davis, Undergraduate, Paul G. Allen School of Computer
Science and Engineering
Former lab members
-
Robin Aguilar,
Scientific and Marketing Communications Specialist, A-Alpha Bio
-
Ferhat
Ay, Institute Leadership Associate Professor of Computational
Biology, La Jolla Institute for Allergy and Immunology
-
Zafer Aydin,
Associate Professor, Computer Enginering Department, Abdullah Gul
University, Kayseri, Turkey
-
Asa Ben-Hur,
Professor, Department of Computer Science, Colorado State University,
Fort Collins
-
Wout Bittremieux,
Assistant Research Professor, Adrem Data Lab, University of Antwerp
-
Giancarlo Bonora, Senior Bioinformatics Scientist, R&D
department, Predicine
-
Xiaoyu Chen, Principal Computational Biologist, Adaptive
Biotechnology
-
Ayse Dincer, Research
Software Engineer, Uber
-
Timothy Durham, Bioinformatics Specialist I, Broad Institute
-
Eleazar Eskin,
Professor, Department of Computer Science, Department of Human
Genetics, University of California, Los Angeles
-
Will Fondrie, Senior Data Scientist, Talus Bioscience
-
Victoria
Haghighi, Professor, Department of Neuroscience,
Icahn School of Medicine at Mount Sinai.
-
Michael Hoffman,
Scientist, Princess Margaret Cancer Centre, Toronto, Canada;
Associate Professor, Department of Medical Biophysics, University of Toronto
-
Borislav Hristov, Assistant Professor, Department of Computer Science,
Cal Poly
-
Lukas Käll,
Professor, Applied Systems Biology, KTH - Royal Institute of
Technology, Sweden
-
Attila
Kertesz-Farkas, Associate Professor, School of Data Analysis and
Artificial Intelligence, the Faculty of Informatics, National Research
University Higher School of Economics in Moscow, Russian
Federation.
-
Aaron Klammer, VP of Software Engineering, Eikon Therapeutics
-
Gang Li,
Co-Principal Investigator, Changping Laboratory
-
Andy Lin, Data
Scientist, Pacific Northwest National Labs
-
Jie
Liu, Assistant Professor, Department of Computational Medicine and
Bioinformatics, University of Michigan
-
Li Liao,
Associate Professor, Department of Computer and Information Sciences,
University of Delaware
-
Max
Libbrecht, Assistant Professor, Department of Computing
Science, Simon Fraser University
-
Yang Lu,
Assistant Professor, Cheriton School of Computer Science, University
of Waterloo
-
Wenxiu Ma, Associate
Professor, Department of Statistics, UC Riverside
-
Damon May,
Computational Immunologist, Adaptive Biotechnologies Corporation
-
Tobias Mann, Director of Software Engineering, Bioinformatics,
Adaptive Biotechnologies Corporation
-
Sean
McIlwain, Assistant Scientist, Department of Biostatistics &
Medical Informatics, University of Wisconsin
-
Alan Min, medical student, University of Washington
-
Merja Oja, VTT Technical Research Centre of Finland
-
Paul Pavlidis,
Professor of Psychiatry, University of British Columbia
-
Lindsay Pino, Co-Founder and
CTO, Talus Bioscience.
-
Sheila
Reynolds, Senior Research Scientist, Institute for Systems
Biology
-
Jacob
Schreiber, Visiting Scientist, Institute of Molecular
Pathology, Vienna.
-
Oliver Serang,
Assistant Professor, Computer Science, Hillsdale College
-
Ritambhara Singh, Associate
Professor, Department of Computer Science, Brown University
-
Ilan Wapinski,
Vice President of Translational Research and Algorithm Products,
PathAI
-
Xiao Wang, Research
Scientist, TikTok
-
Gurkan
Yardimci, Assistant Professor, Oncological Sciences Division,
Oregon Health Sciences University
-
Melih Yilmaz,
Applied Scientist, Amazon
-
Habil Zare, Assistant
Professor, Department of Cell Systems & Anatomy, University of Texas
Health Science Center at San Antonio
-
Ran Zhang, Assistant
Professor, School of Data Science and Society, University of North
Carolina Chapel Hill
|